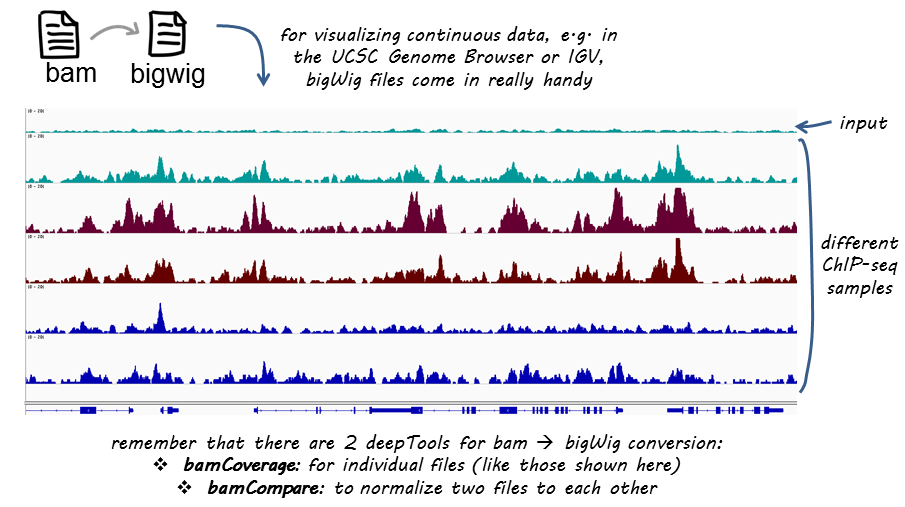
Podstawy komputerowej analizy danych pochodzących z ChIP-seq :: Biotechnologia: e-biotechnologia.pl :: Biotechnologiczny Portal Internetowy-aktualności, artykuły, laboratorium, studia biotechnologiczne.
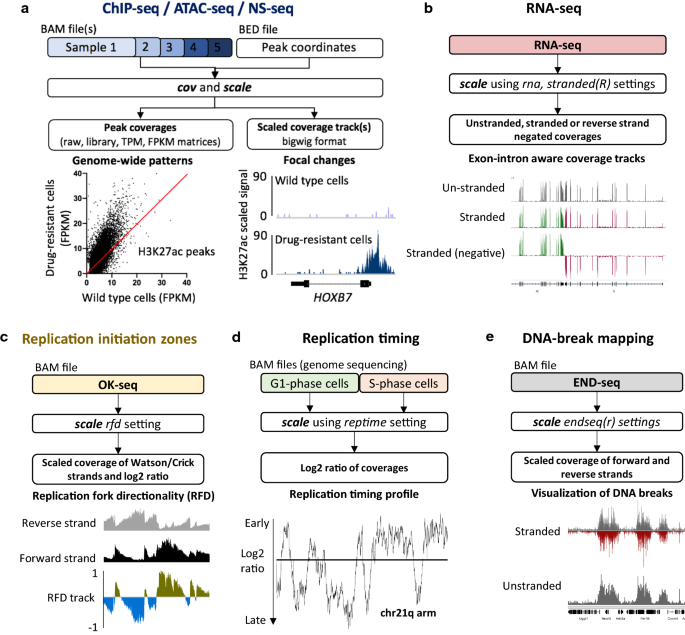
BAMscale: quantification of next-generation sequencing peaks and generation of scaled coverage tracks | Epigenetics & Chromatin | Full Text



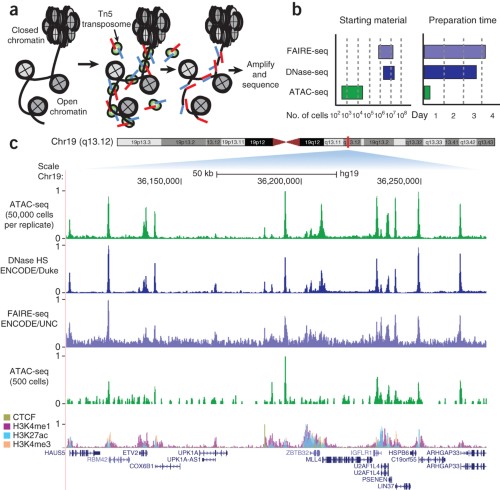
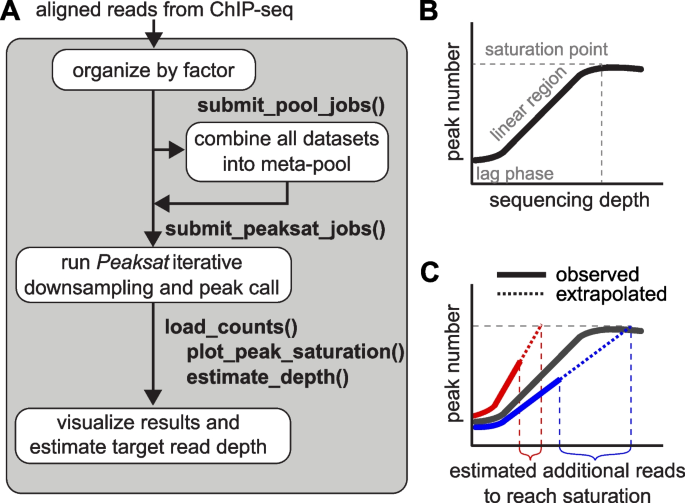
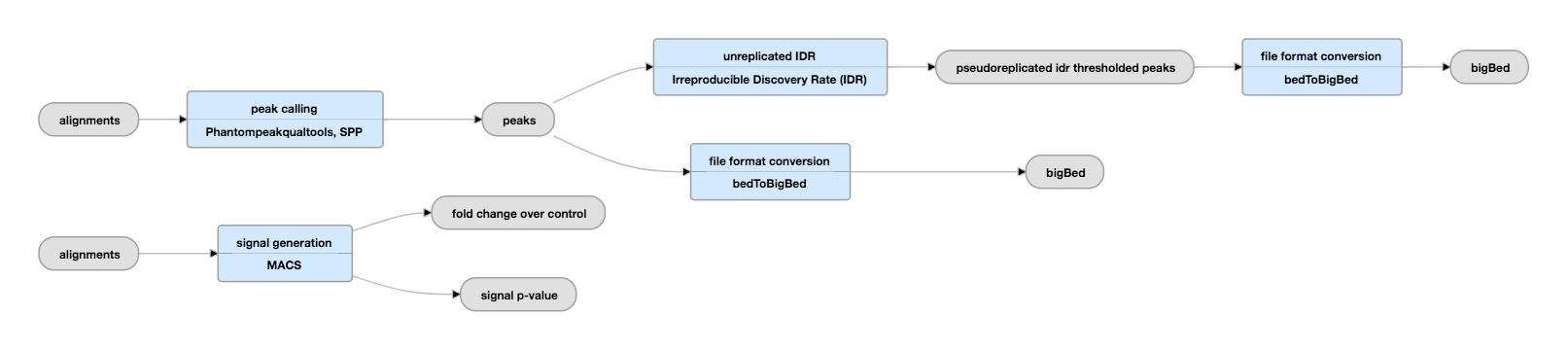
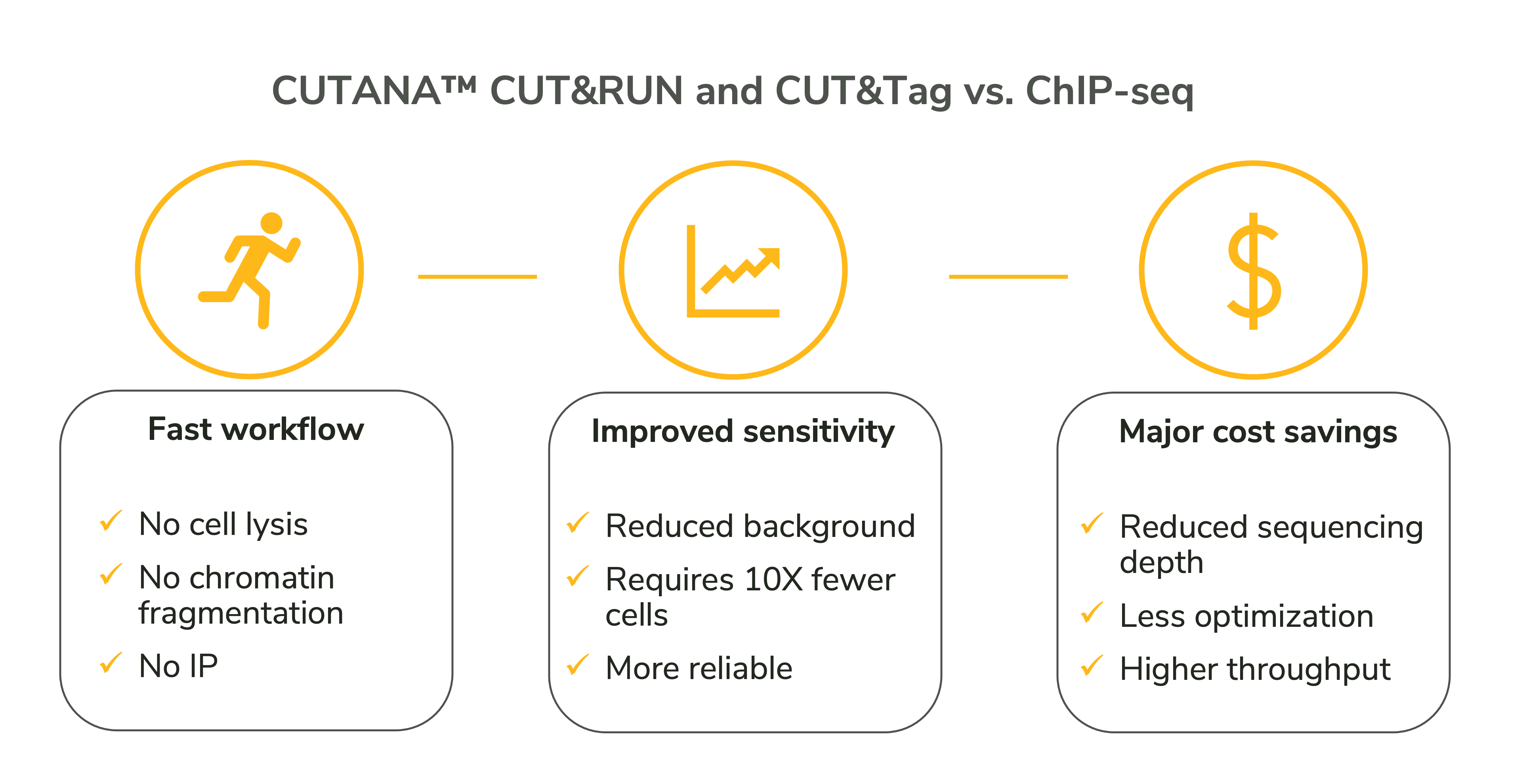
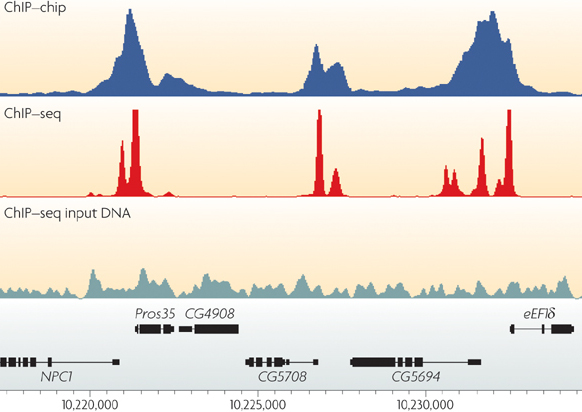
![PDF] Practical Guidelines for the Comprehensive Analysis of ChIP-seq Data | Semantic Scholar PDF] Practical Guidelines for the Comprehensive Analysis of ChIP-seq Data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/0e1a208d7ef393571c1e0221839737d964bd1f3f/2-Figure1-1.png)