Advances of DNase-seq for mapping active gene regulatory elements across the genome in animals - ScienceDirect

Nature Reviews Genetics on Twitter: "Principal methods for measuring chromatin accessibility: #DNAse-seq #ATAC-seq #MNase-seq #NOMe-seq Figure from our Review: Chromatin accessibility and the regulatory epigenome https://t.co/SYwpxOCv81 by @WJGreenleaf ...
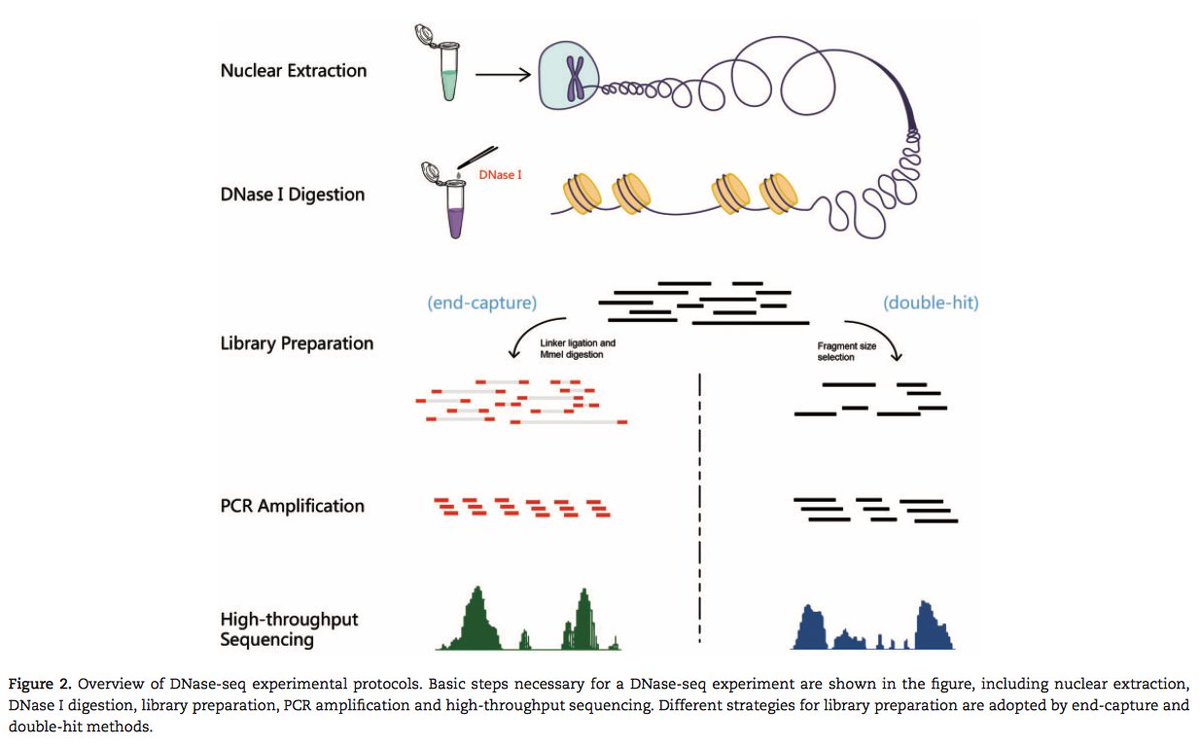
DNase-seq: A High-Resolution Technique for Mapping Active Gene Regulatory Elements across the Genome from Mammalian Cells

DNase Seq | DNase Footprinting | DNase1 Footprinting | DNase 1 Hypersensitive Sites Sequencing | - YouTube
![PDF] Explicit DNase sequence bias modeling enables high-resolution transcription factor footprint detection | Semantic Scholar PDF] Explicit DNase sequence bias modeling enables high-resolution transcription factor footprint detection | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/800d7add4aada32c5e80b1e1cf6cc33319828da3/5-Figure1-1.png)
PDF] Explicit DNase sequence bias modeling enables high-resolution transcription factor footprint detection | Semantic Scholar
Ensembl on Twitter: "#Method paper: Practical guide for #DNase-seq #analysis for #chromatin accessibility, from #datamanagement to #dataviz. Summarising and explaining available tools & experimental protocols. #CitedEnsembl #bioinformatics #phdchat ...

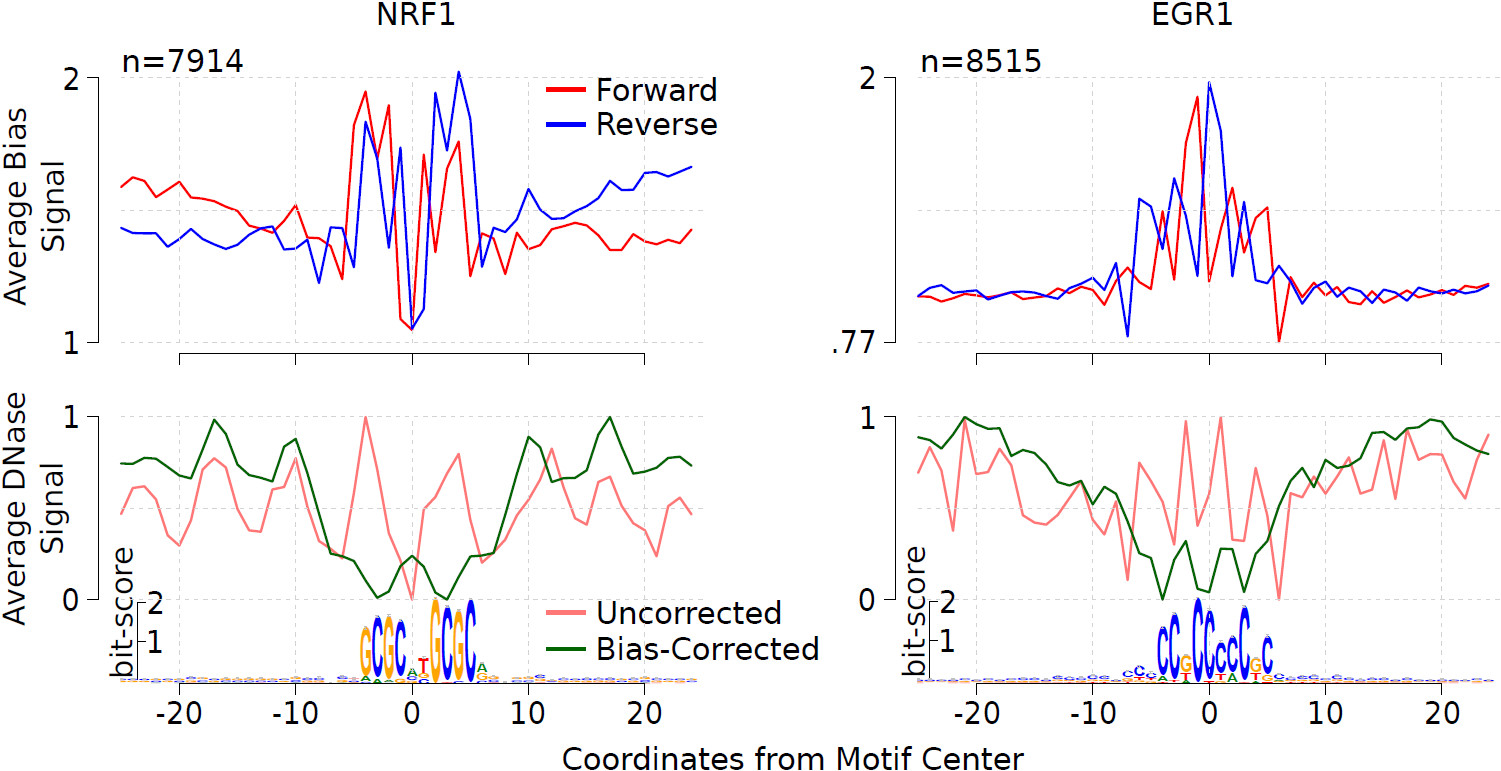
Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification | Nature Methods
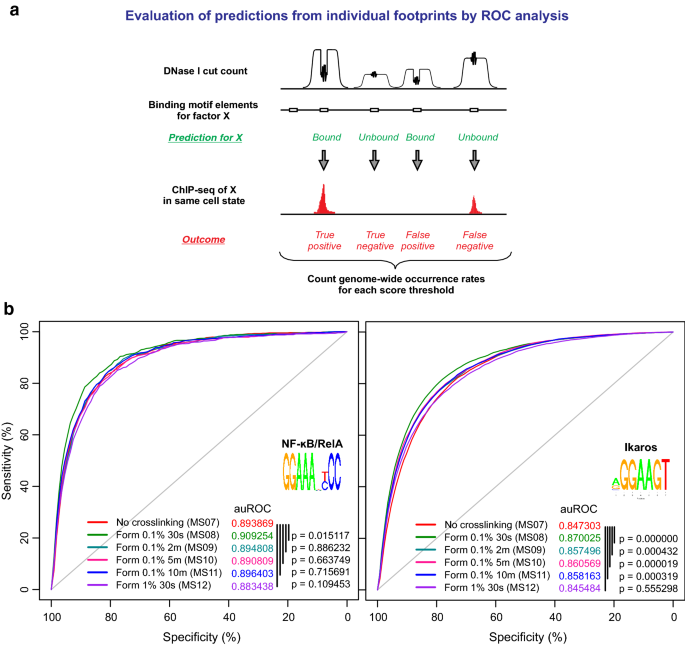
XL-DNase-seq: improved footprinting of dynamic transcription factors | Epigenetics & Chromatin | Full Text

DNase I hypersensitivity mapping, genomic footprinting, and transcription factor networks in plants - ScienceDirect
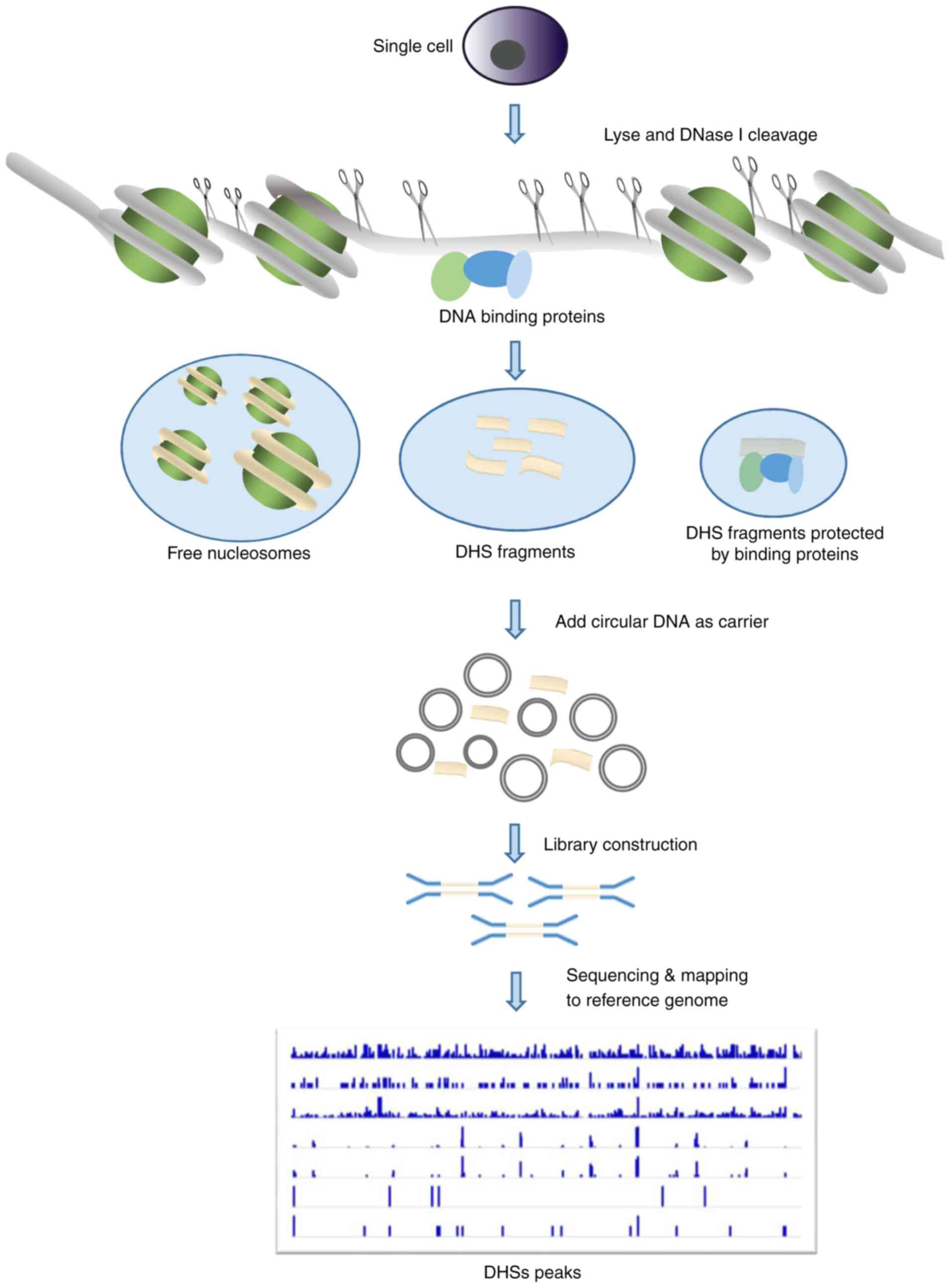
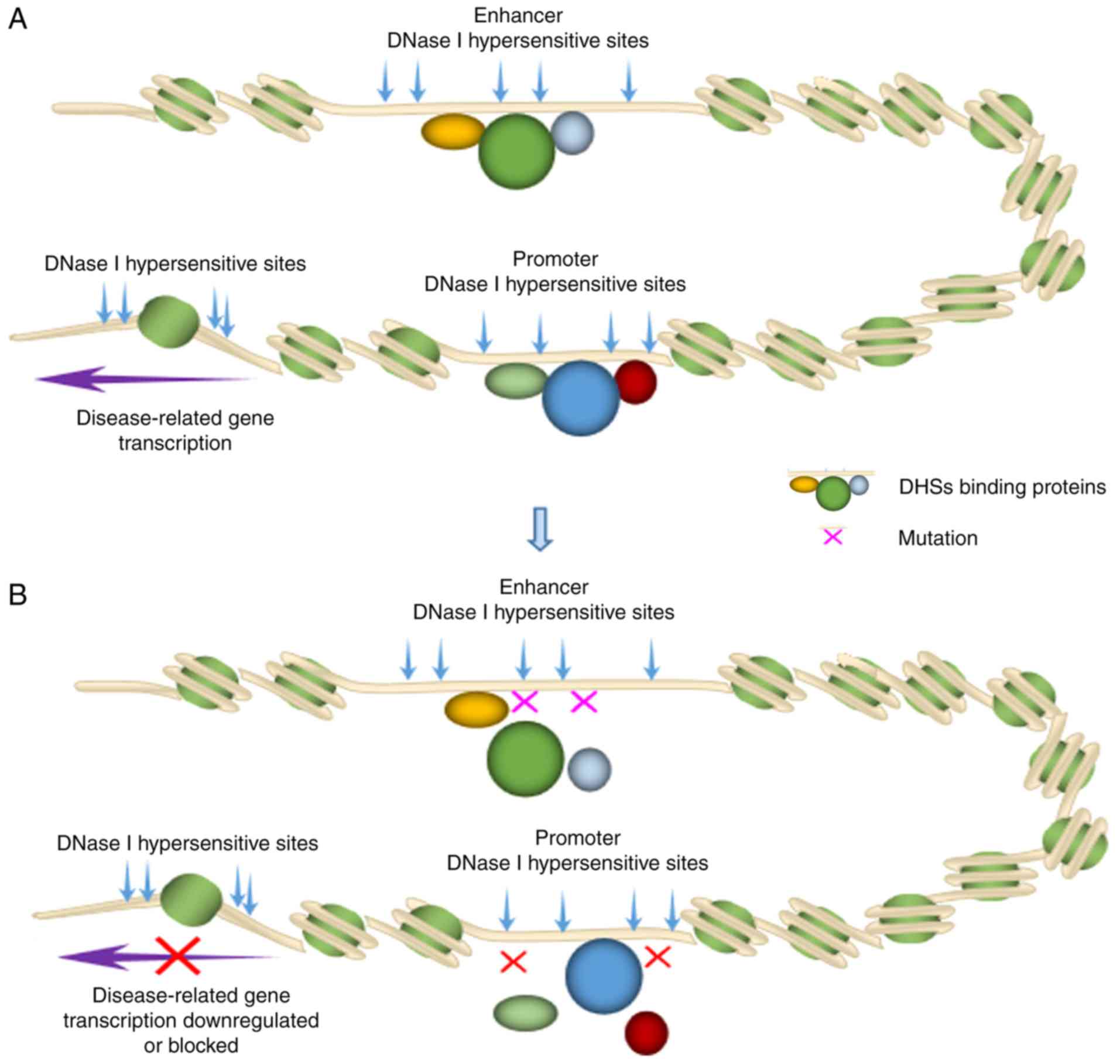
Unveiling the gene regulatory landscape in diseases through the identification of DNase I‑hypersensitive sites (Review)

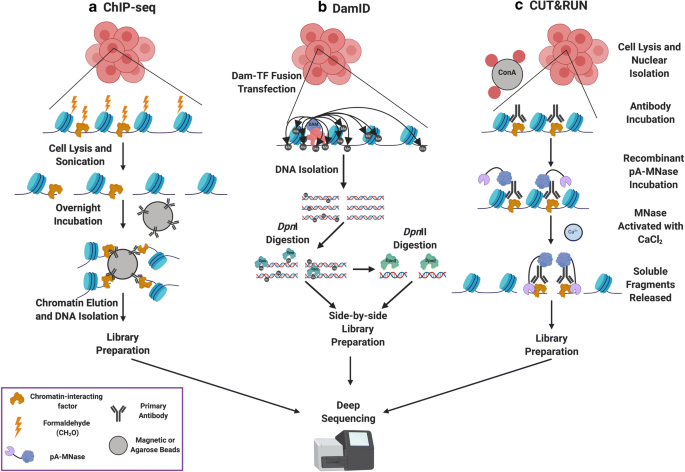
IJMS | Free Full-Text | Emerging Single-Cell Technological Approaches to Investigate Chromatin Dynamics and Centromere Regulation in Human Health and Disease