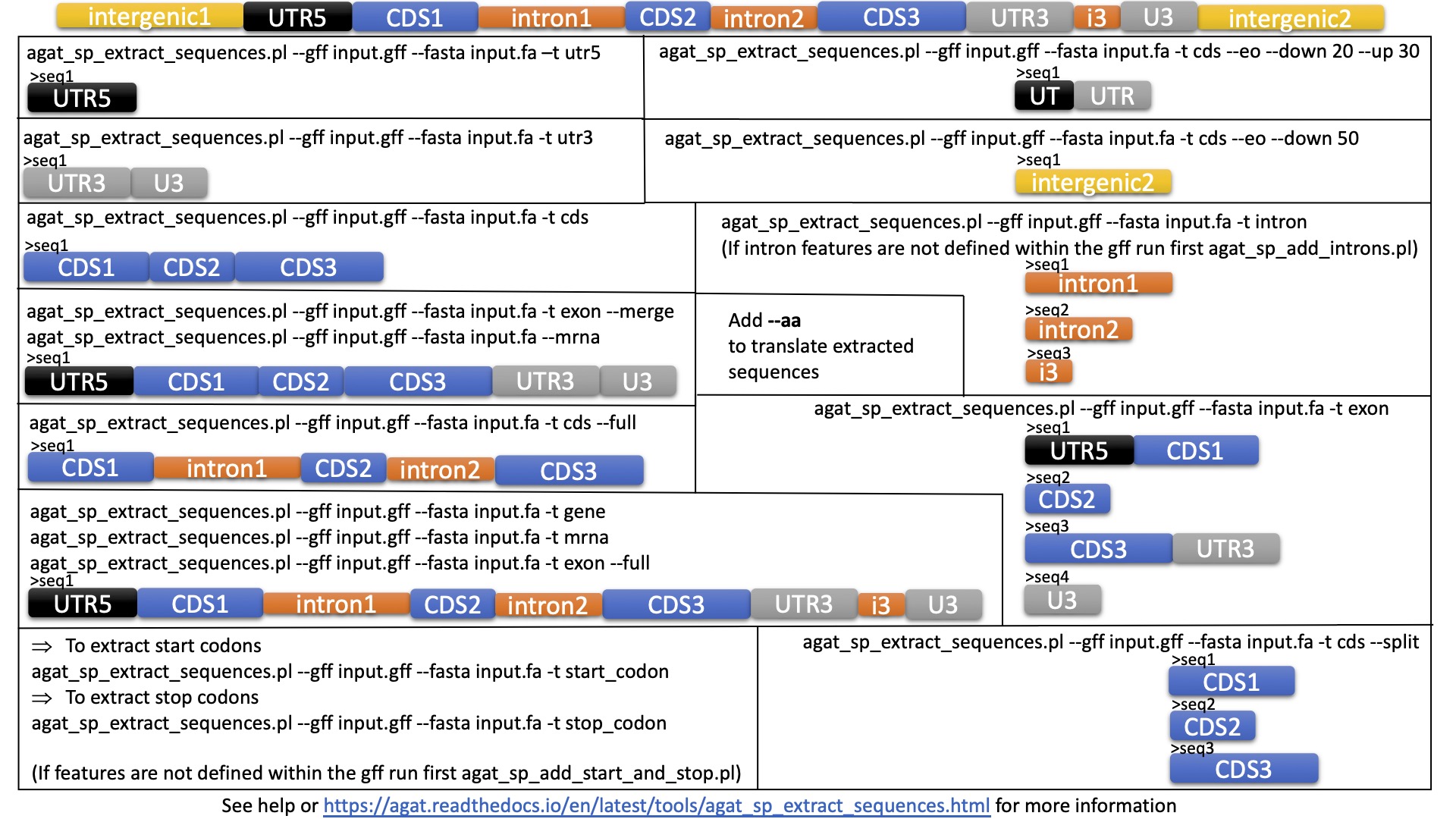
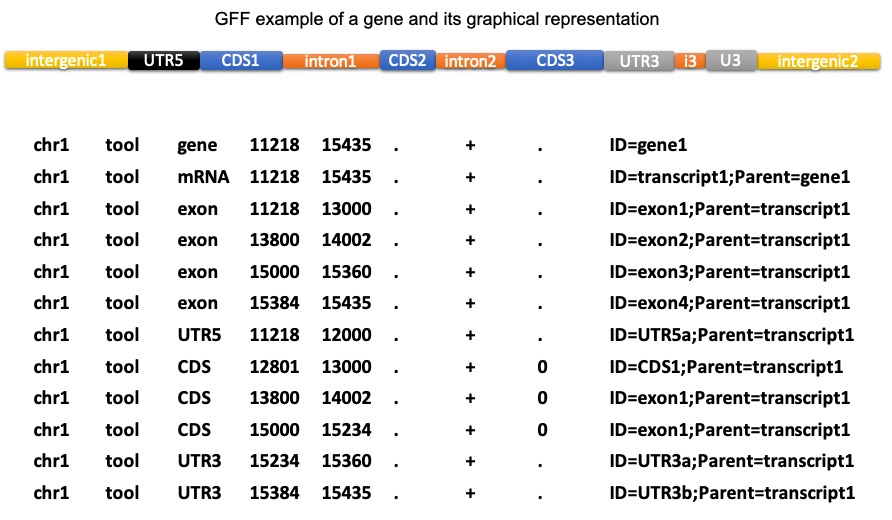
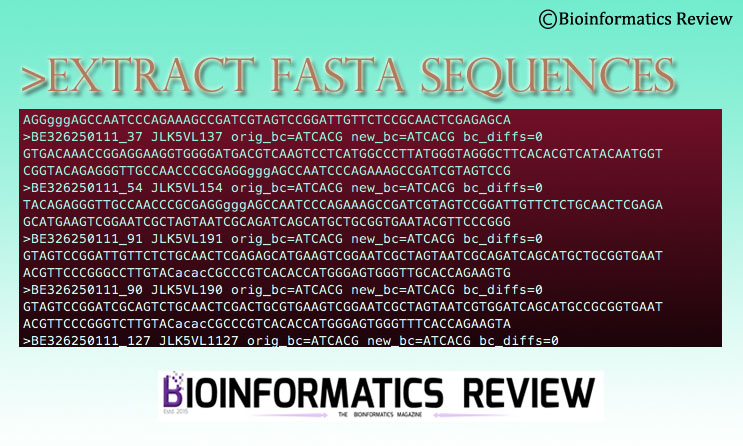
How to extract fasta sequences from a multi-fasta file based on matching headers in a separate file? — Bioinformatics Review
GitHub - IdoBar/FastRetriever: A python script used to extract specific sequences from a fasta file based on ID or keyword
How to extract fasta seq using gene id and not transcript id from stringtie output (gene count matrix file) · Issue #6 · gpertea/gffread · GitHub