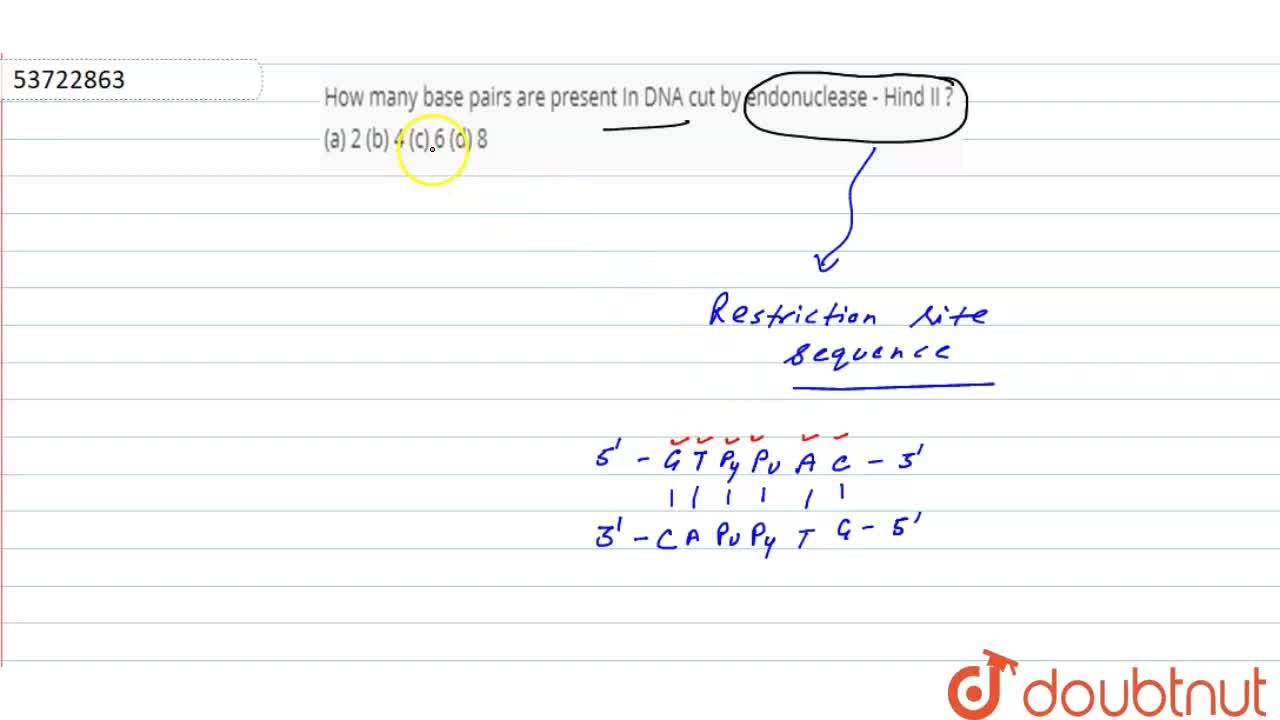
Hind-II always cut DNA molecules at a particular point by recognising a specific sequence of six - YouTube

It was found that Hind 2 always cuts DNA molecules at a particular point by recognising a specific sequence - Brainly.in

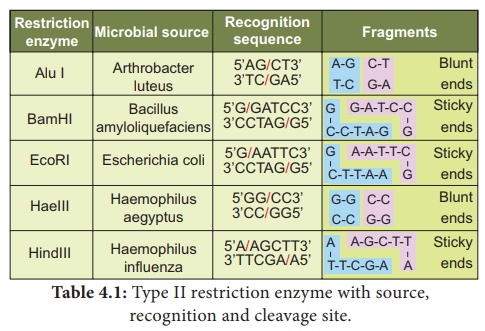
What is the Difference Between EcoRI and HindIII Restriction Enzymes | Compare the Difference Between Similar Terms

SOLVED: 'If the recognition sequence of the restriction enzyme Hindlll is AAGCTT, then how many covalent bonds will be broken by the enzyme in the following DNA molecule? STCAAGCTCC-G-A-A-G-CTTGA JAGTSTCGAAGCTTC-GAACT 5 8.2
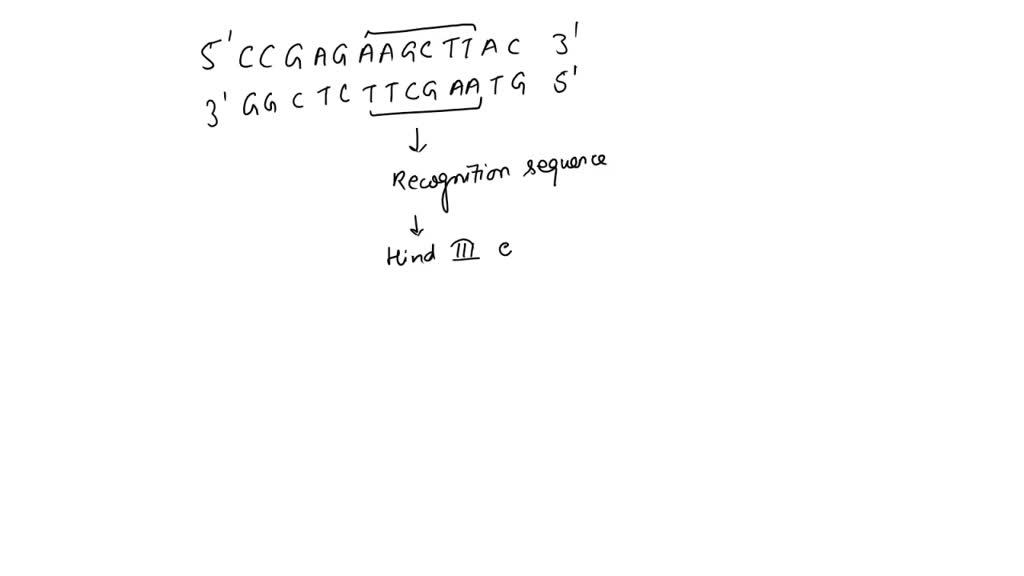
SOLVED: The DNA sequence (5 '-CCGAGAGATCTAC-3') contains a six-base sequence that is a recognition and cutting site for a restriction enzyme. ( 4 points) (a) What is this sequence? (2 points) (b)
Genetic Engineering Info: The nomenclature of restriction enzymes uses a three letter in DNA: E. coli, HpaII , MspI

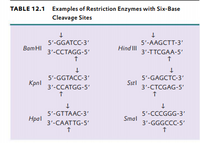
The restriction enzyme responsible for the cleavage of following sequence is 5' - G - T - C - G - A - C - 3'3' - C - A - G - C - T - G - 5'

The restriction enzyme responsible for the cleavage of following sequence is 5' - G - T - C - G - A - C - 3'3' - C - A - G - C - T - G - 5'