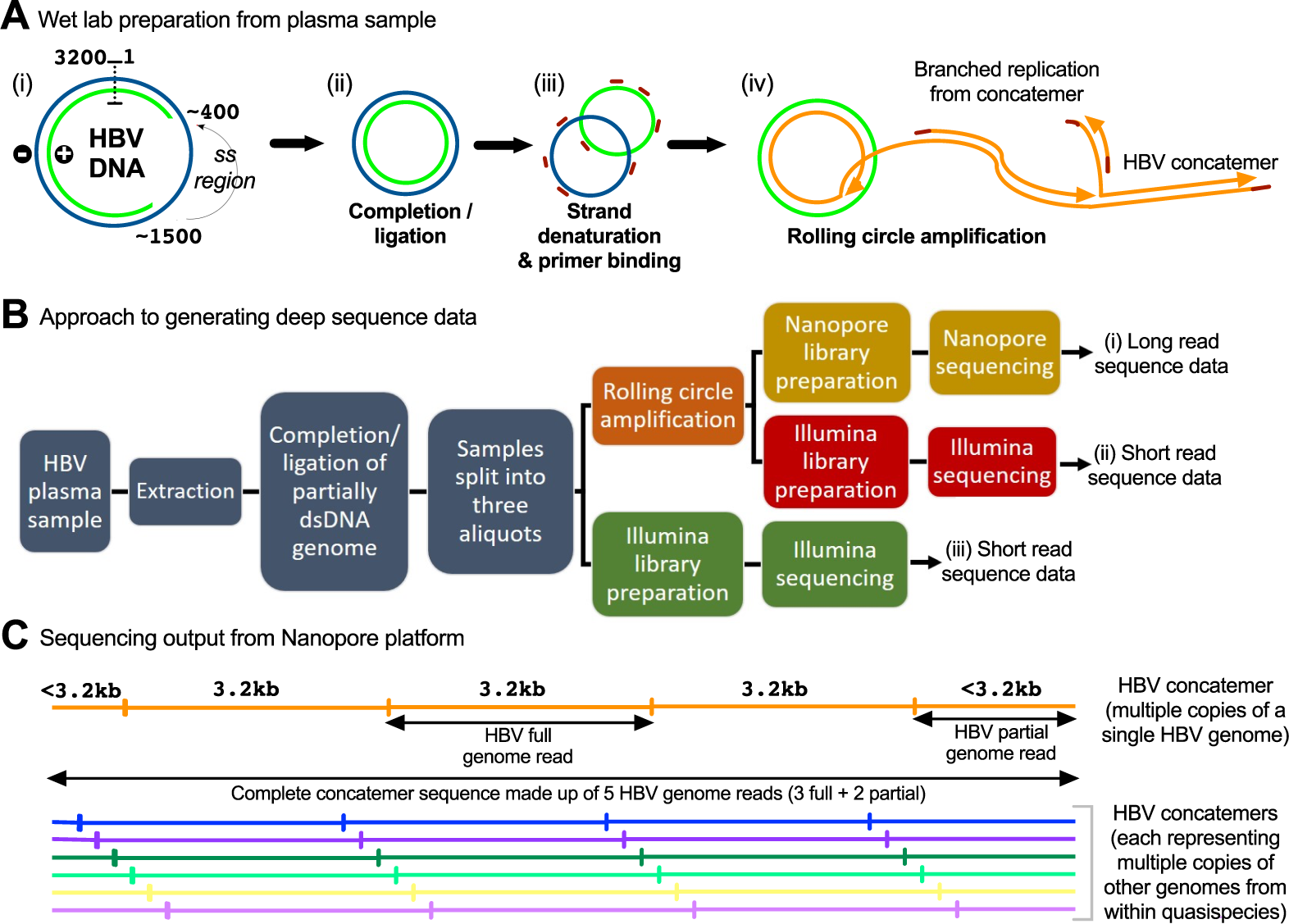
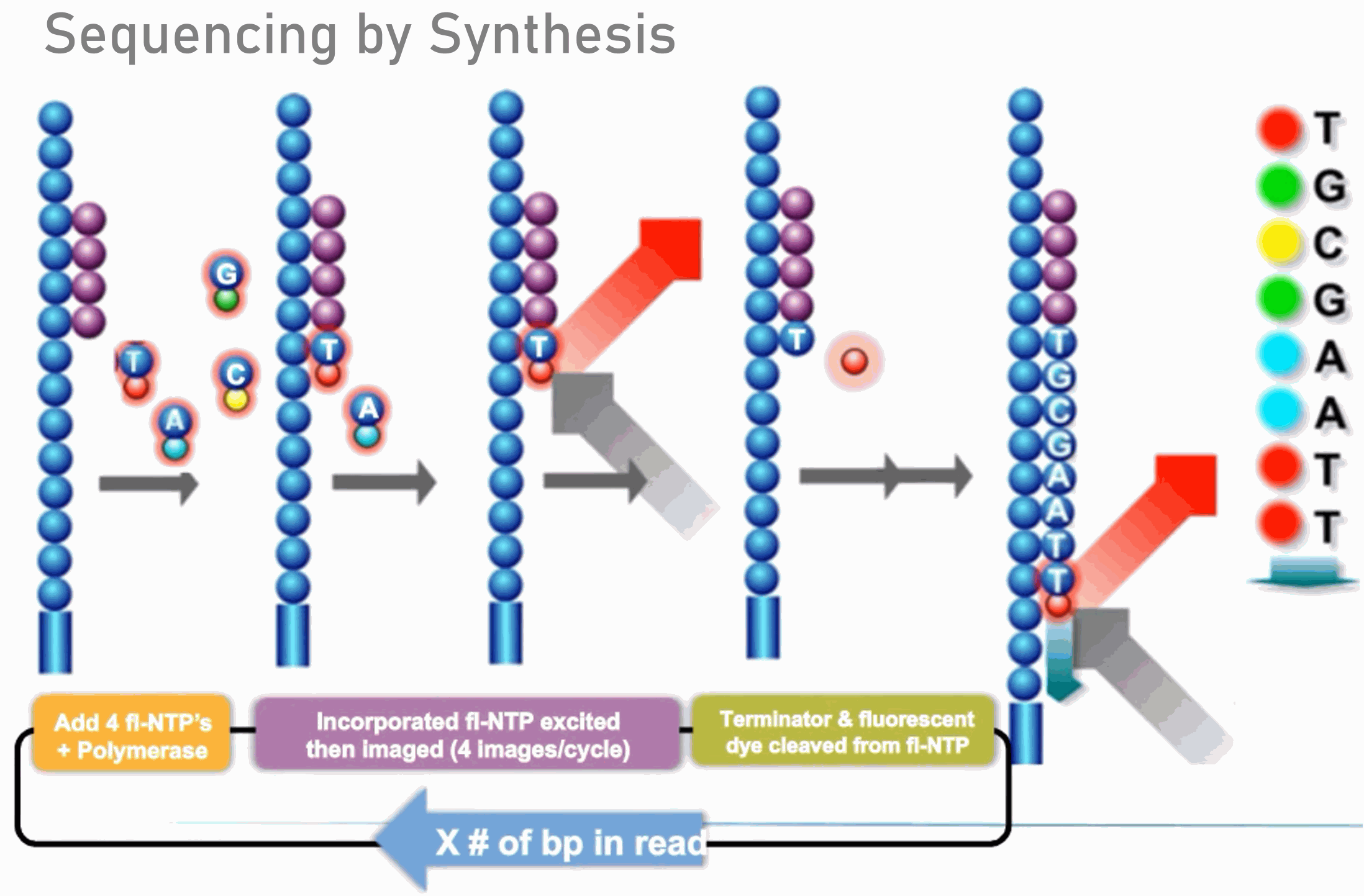
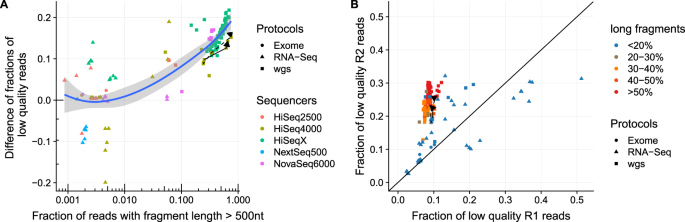
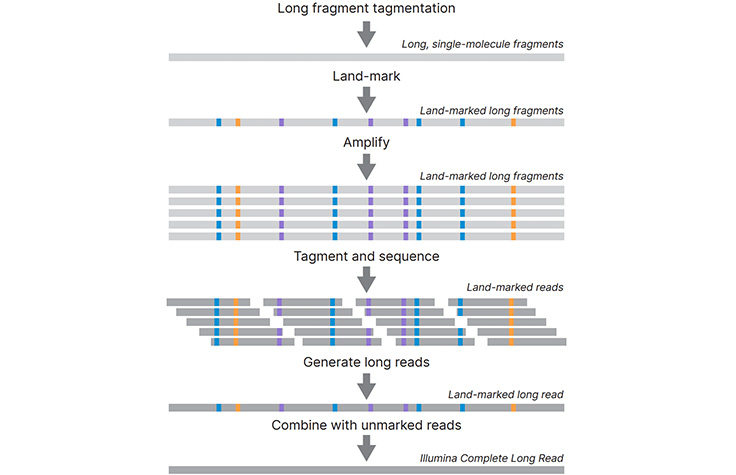
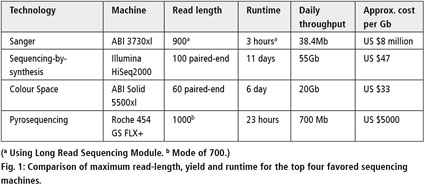
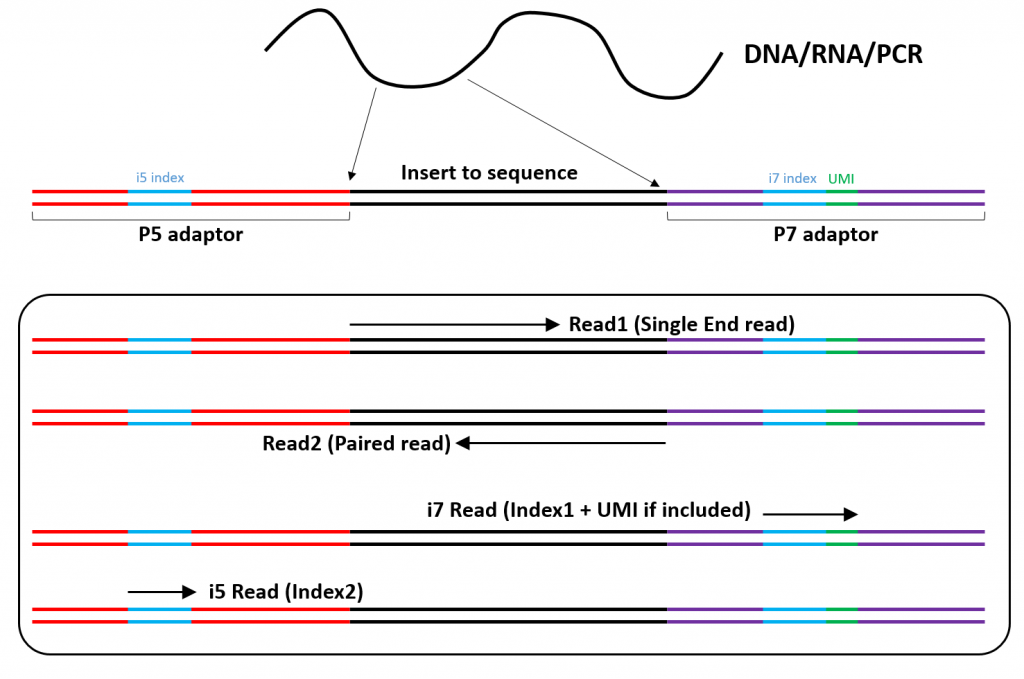
Adapter trimming: Why are adapter sequences trimmed from only the 3' ends of reads - Illumina Knowledge
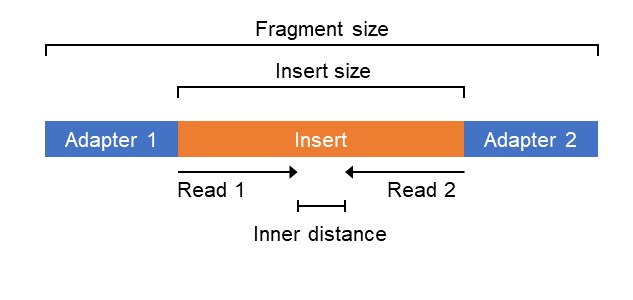
Compbio 020: Reads, fragments and inserts - what you need to know for understanding your sequencing data — Bad Grammar, Good Syntax
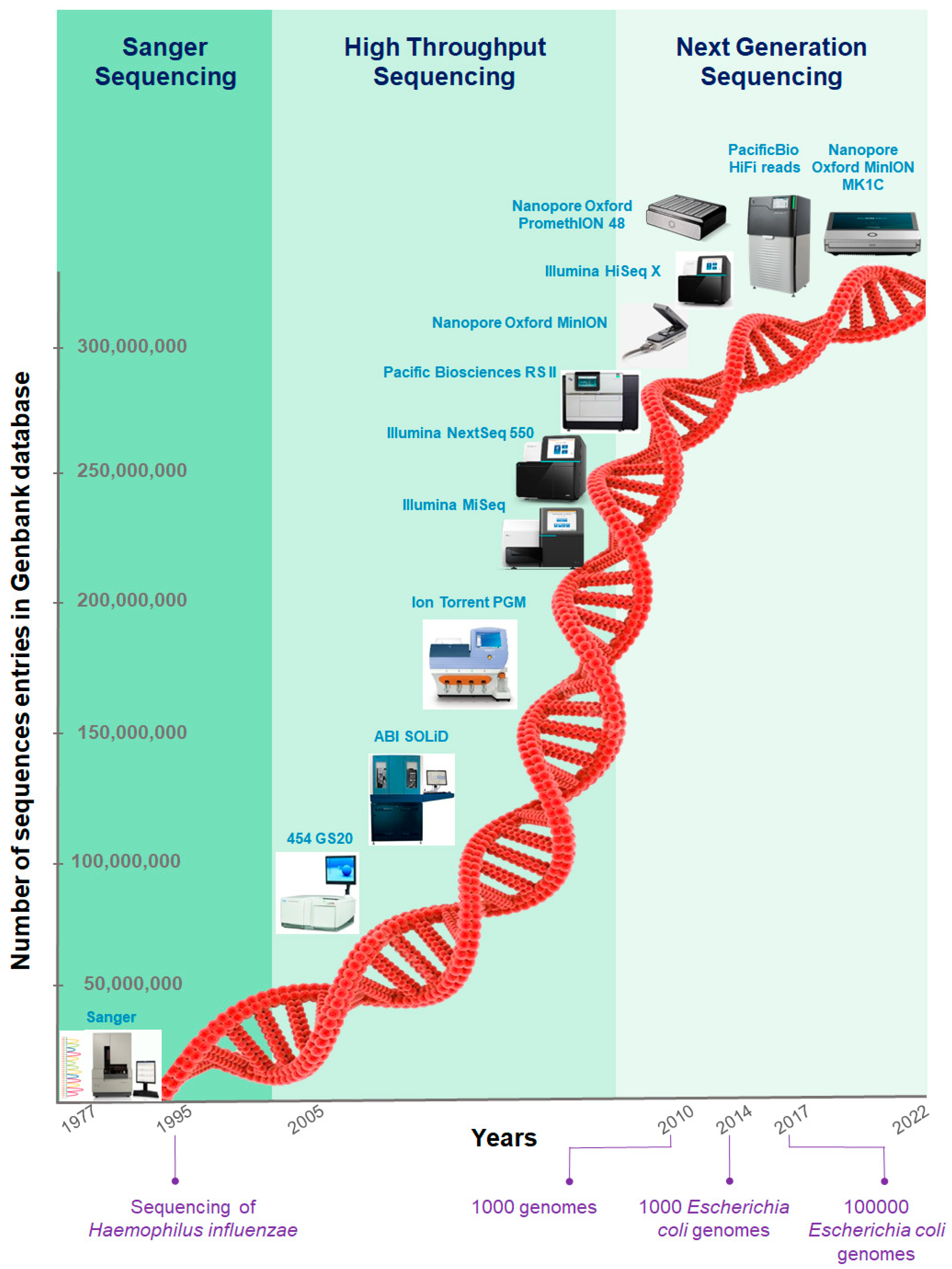
IJMS | Free Full-Text | Application and Challenge of 3rd Generation Sequencing for Clinical Bacterial Studies

The distribution of read lengths in 454, Illumina and PacBio budgerigar... | Download Scientific Diagram
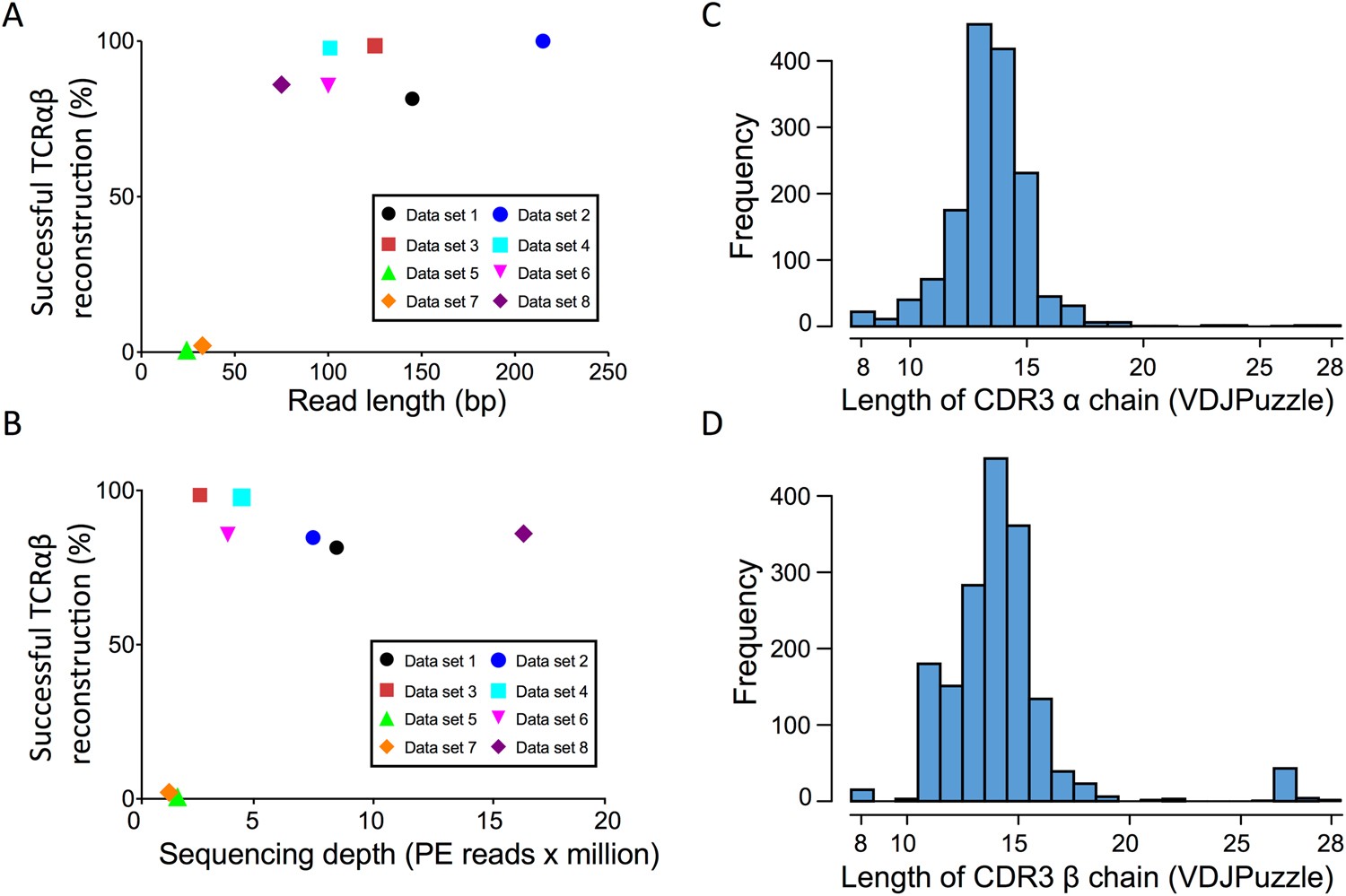
Impact of sequencing depth and read length on single cell RNA sequencing data of T cells | Scientific Reports

Histogram of read variants' lengths in Illumina sequencing data. The... | Download Scientific Diagram