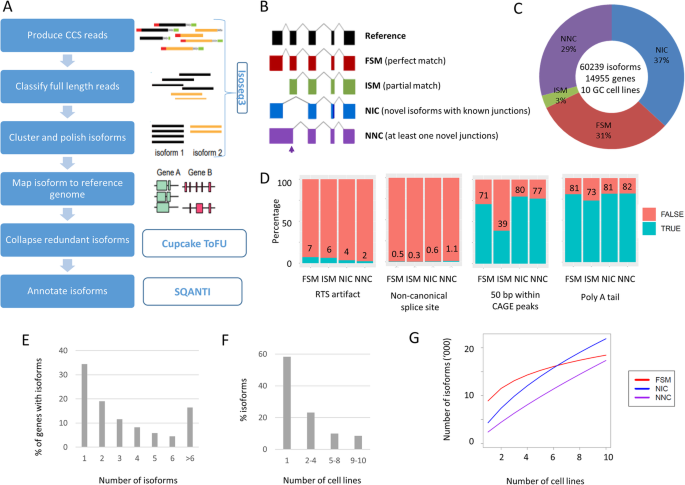
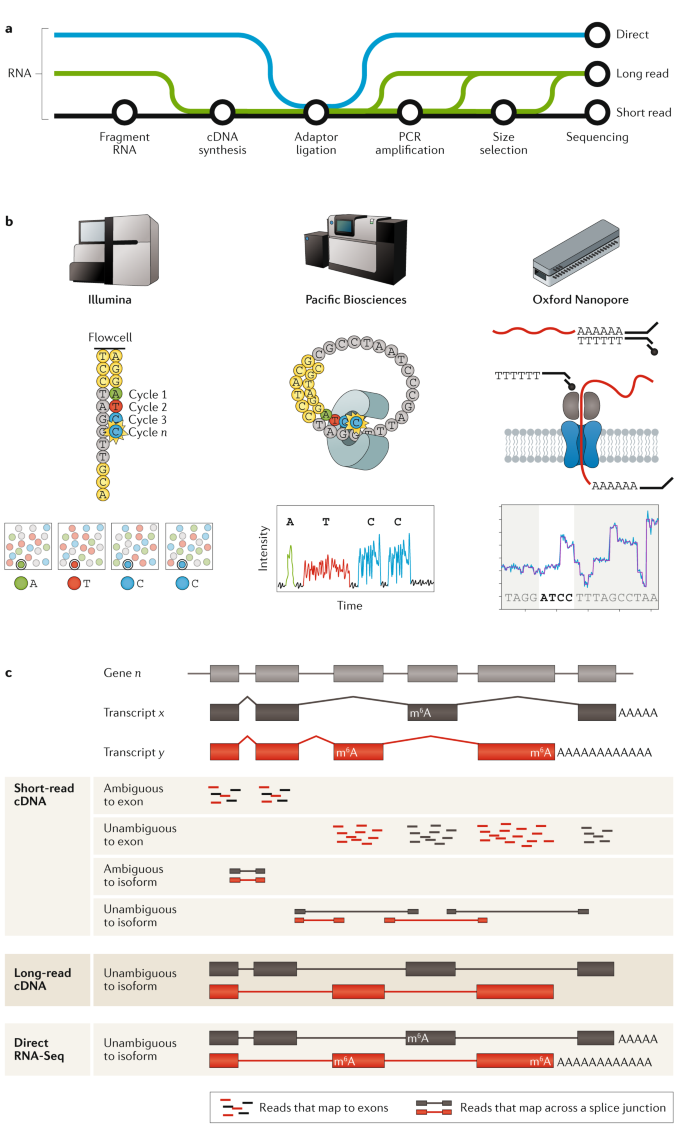
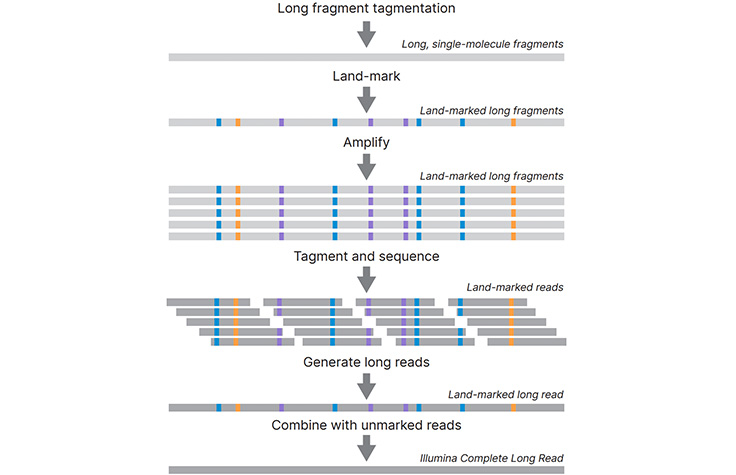
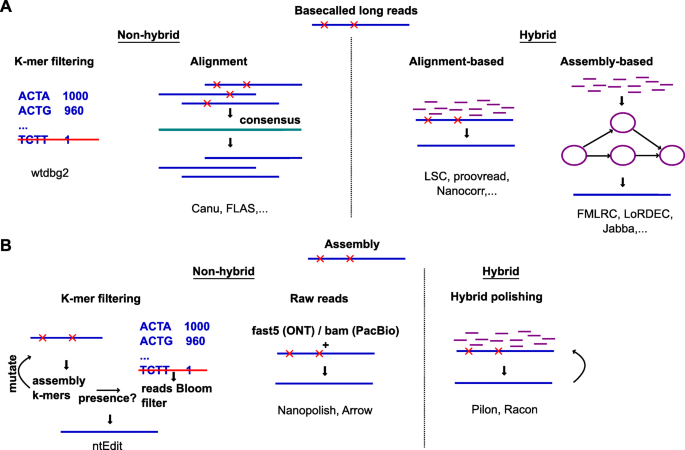
A technology-agnostic long-read analysis pipeline for transcriptome discovery and quantification | bioRxiv

Full Length Isoform Sequencing (Iso-Seq) Yields a More Comprehensive View of Gene Activity - Drug Discovery World (DDW)
Single-molecule long-read sequencing reveals a conserved intact long RNA profile in sperm | Nature Communications

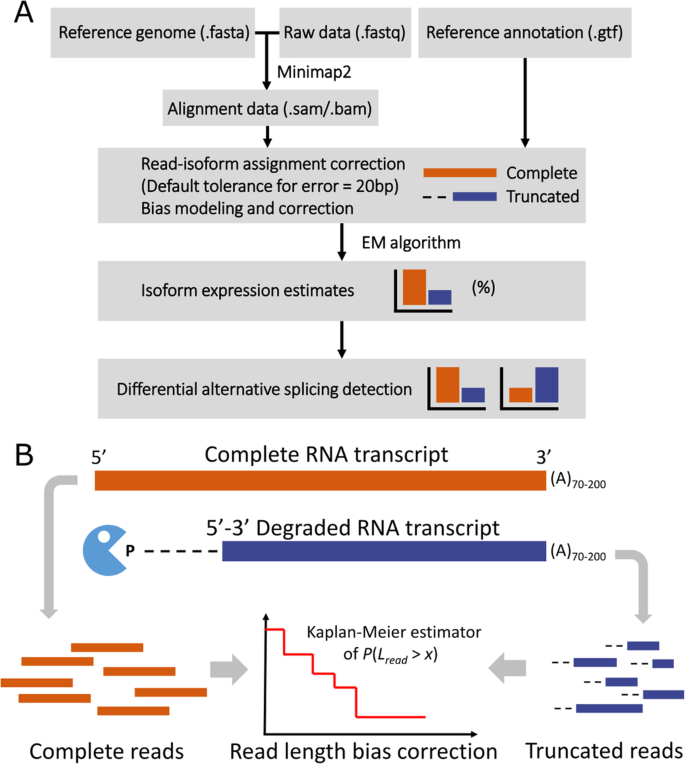
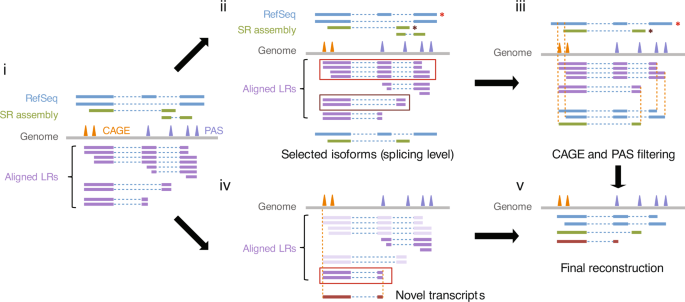
ESPRESSO: Robust discovery and quantification of transcript isoforms from error-prone long-read RNA-seq data | Science Advances

IJMS | Free Full-Text | L-RAPiT: A Cloud-Based Computing Pipeline for the Analysis of Long-Read RNA Sequencing Data

Full-length transcript sequencing of human and mouse cerebral cortex identifies widespread isoform diversity and alternative splicing - ScienceDirect

Nanopore long-read RNA-seq and absolute quantification delineate transcription dynamics in early embryo development of an insect pest | Scientific Reports

A systematic benchmark of Nanopore long read RNA sequencing for transcript level analysis in human cell lines | bioRxiv
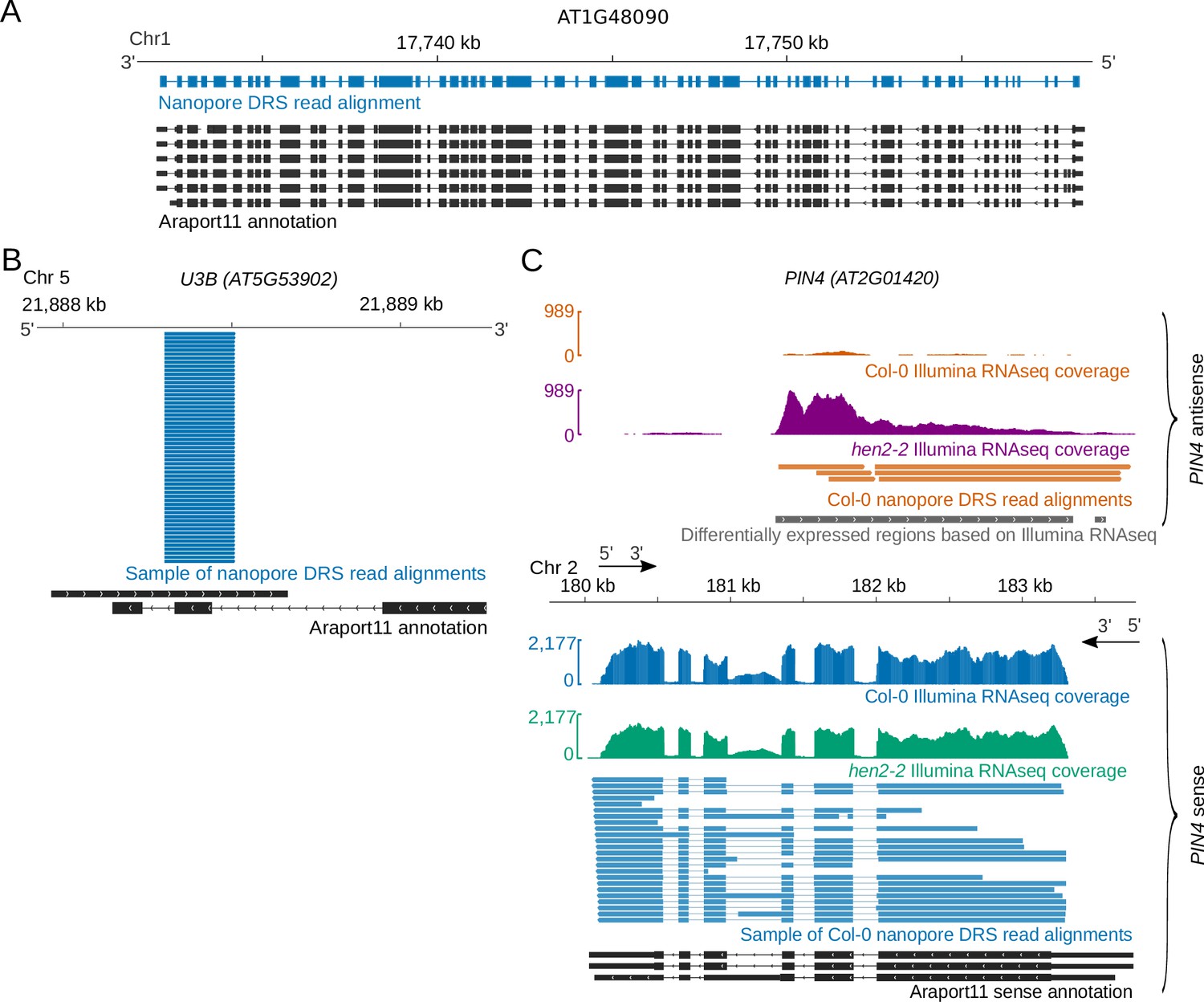
Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification | eLife

Researchers use a combination of third generation long-read RNA sequencing technology and short-read RNAseq to characterize the splicing landscape of prostate cancer | RNA-Seq Blog