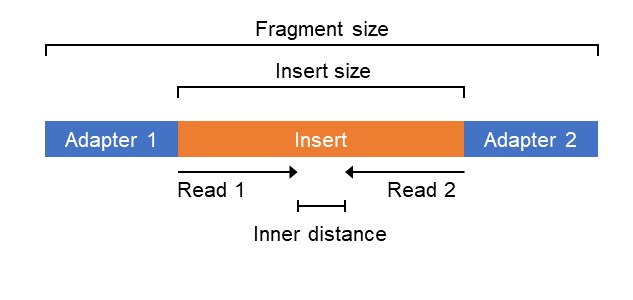
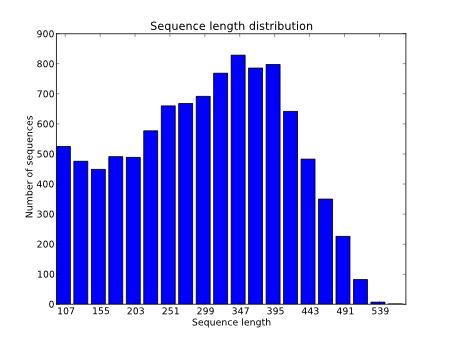
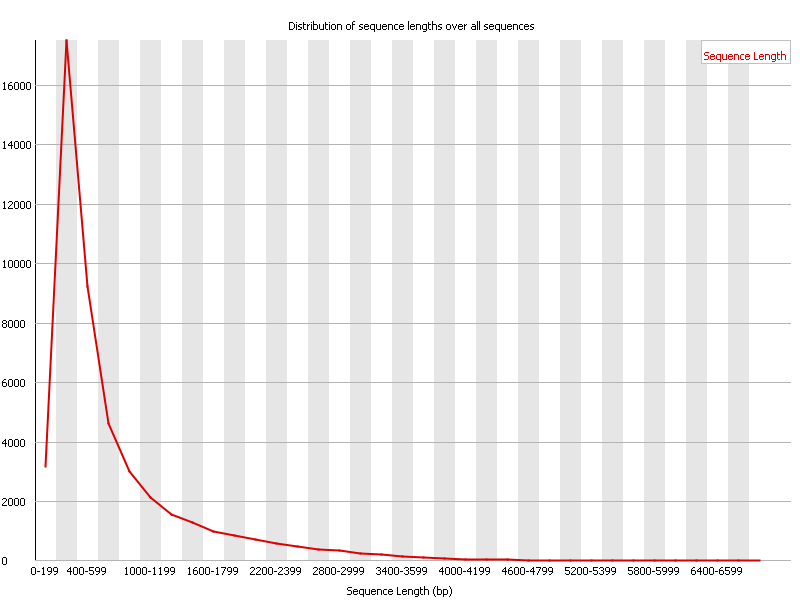
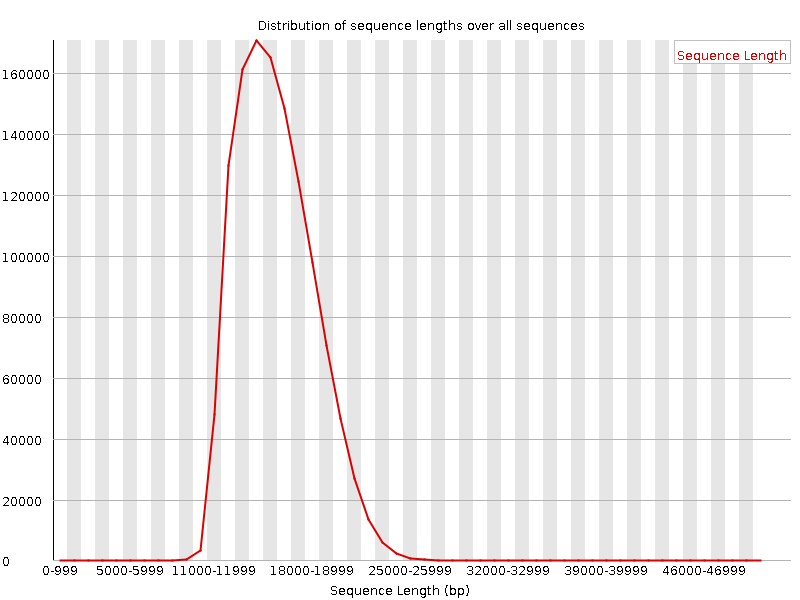
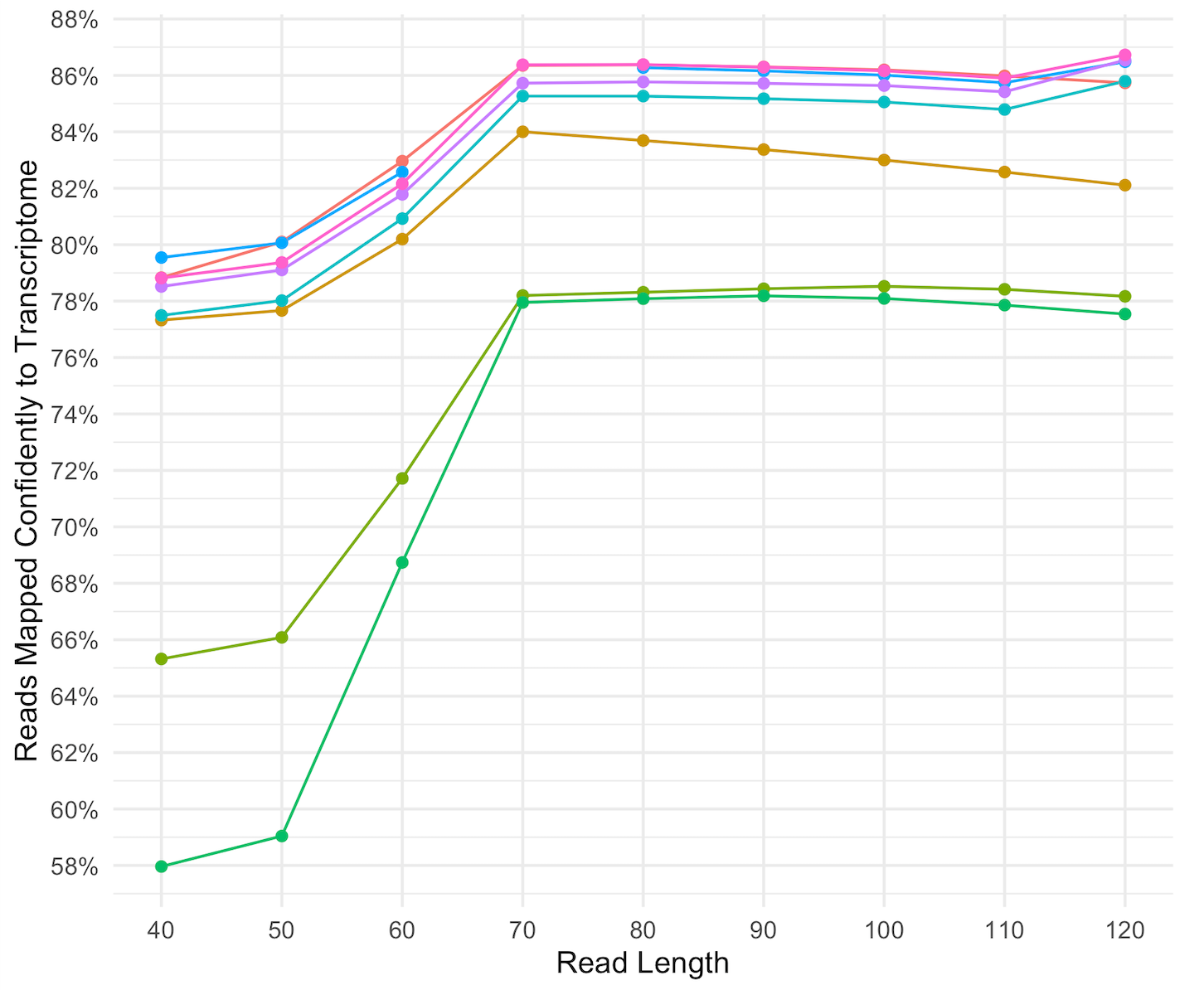
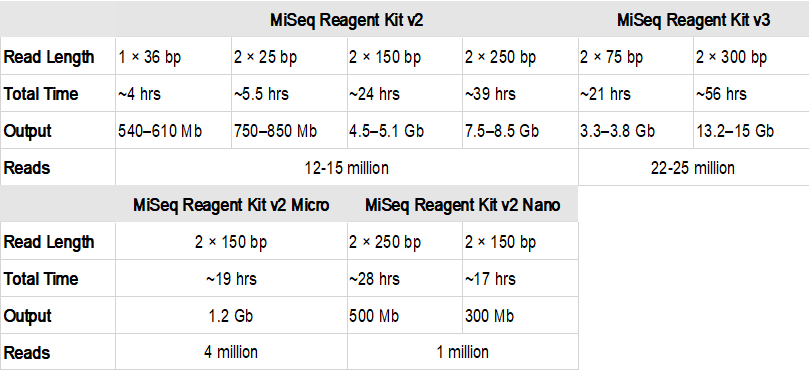
Unusual difference between "Demultiplexed sequence length summary" and " Sequencing company: read length" - User Support - QIIME 2 Forum
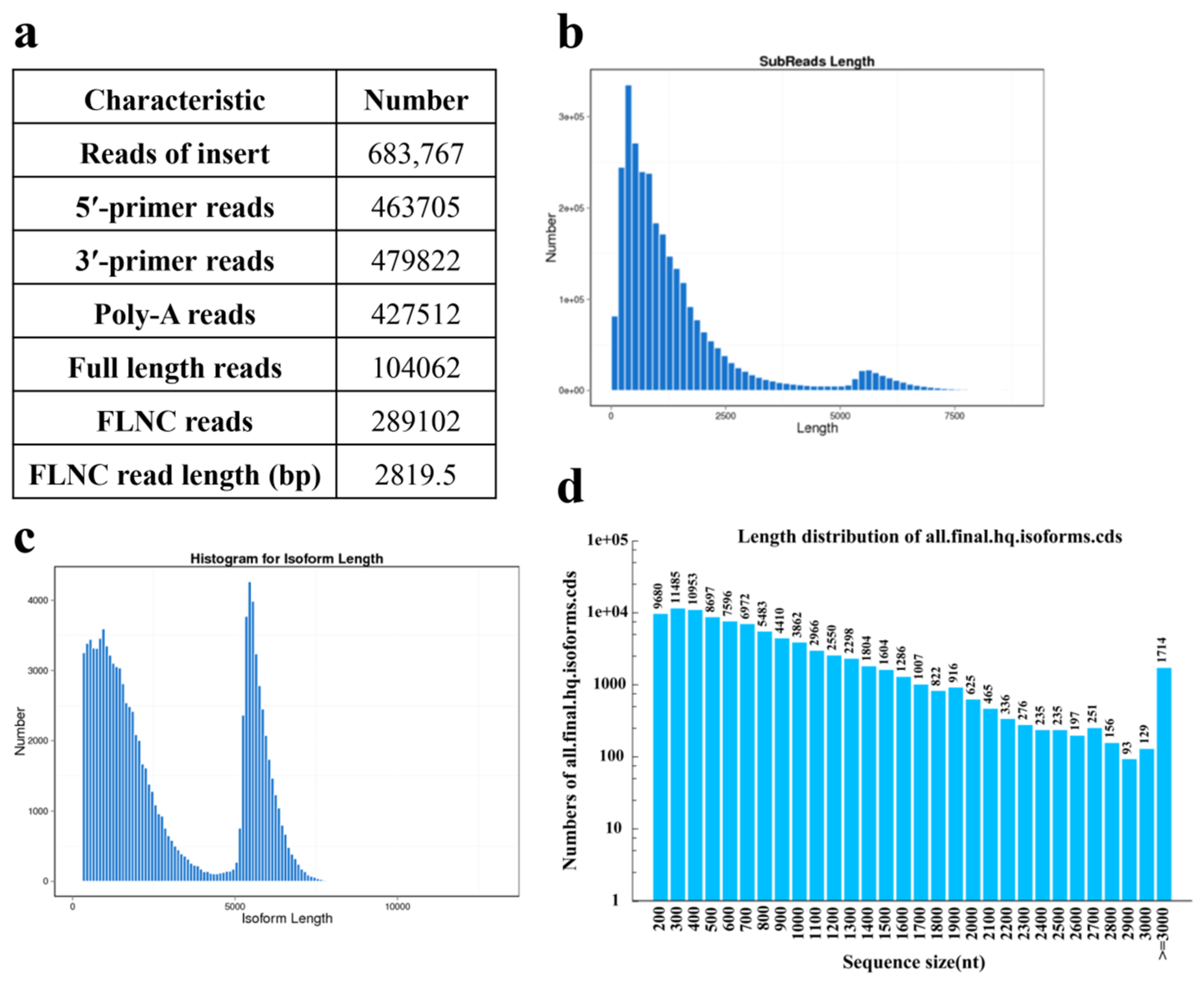
IJMS | Free Full-Text | Full-Length Transcriptome Sequencing and Different Chemotype Expression Profile Analysis of Genes Related to Monoterpenoid Biosynthesis in Cinnamomum porrectum

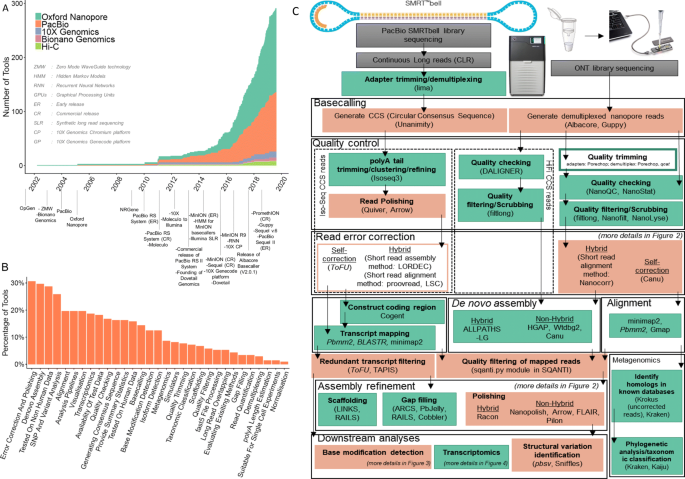
Oxford Nanopore Technology: A Promising Long-Read Sequencing Platform To Study Exon Connectivity and Characterize Isoforms of Complex Genes | Semantic Scholar

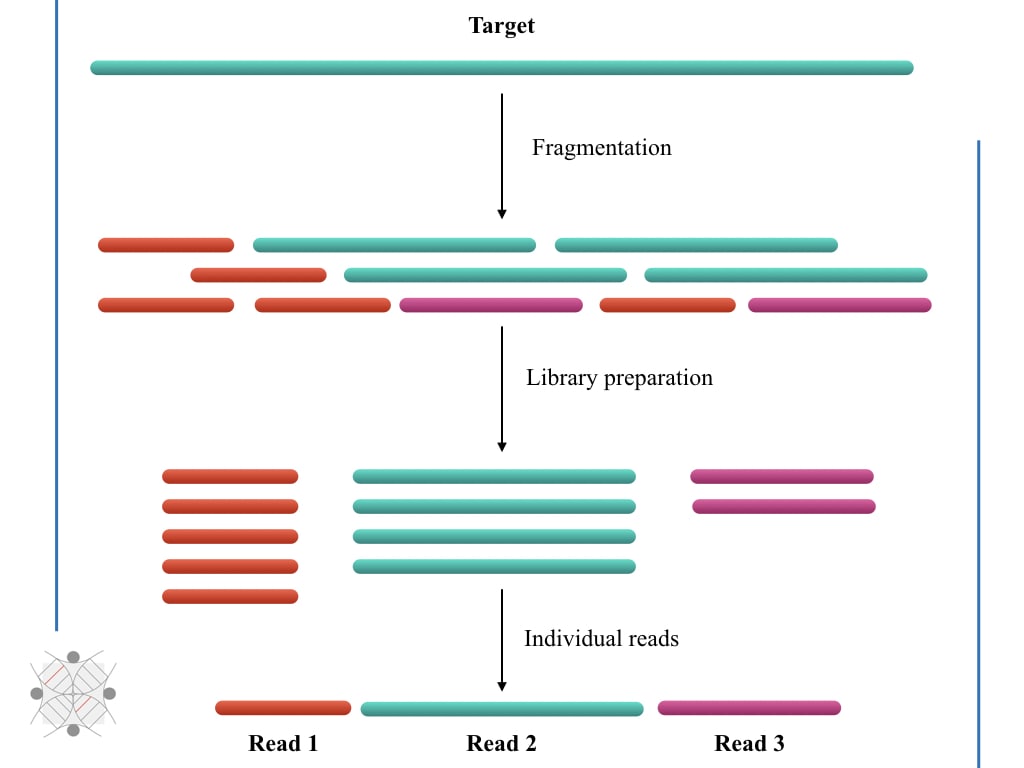
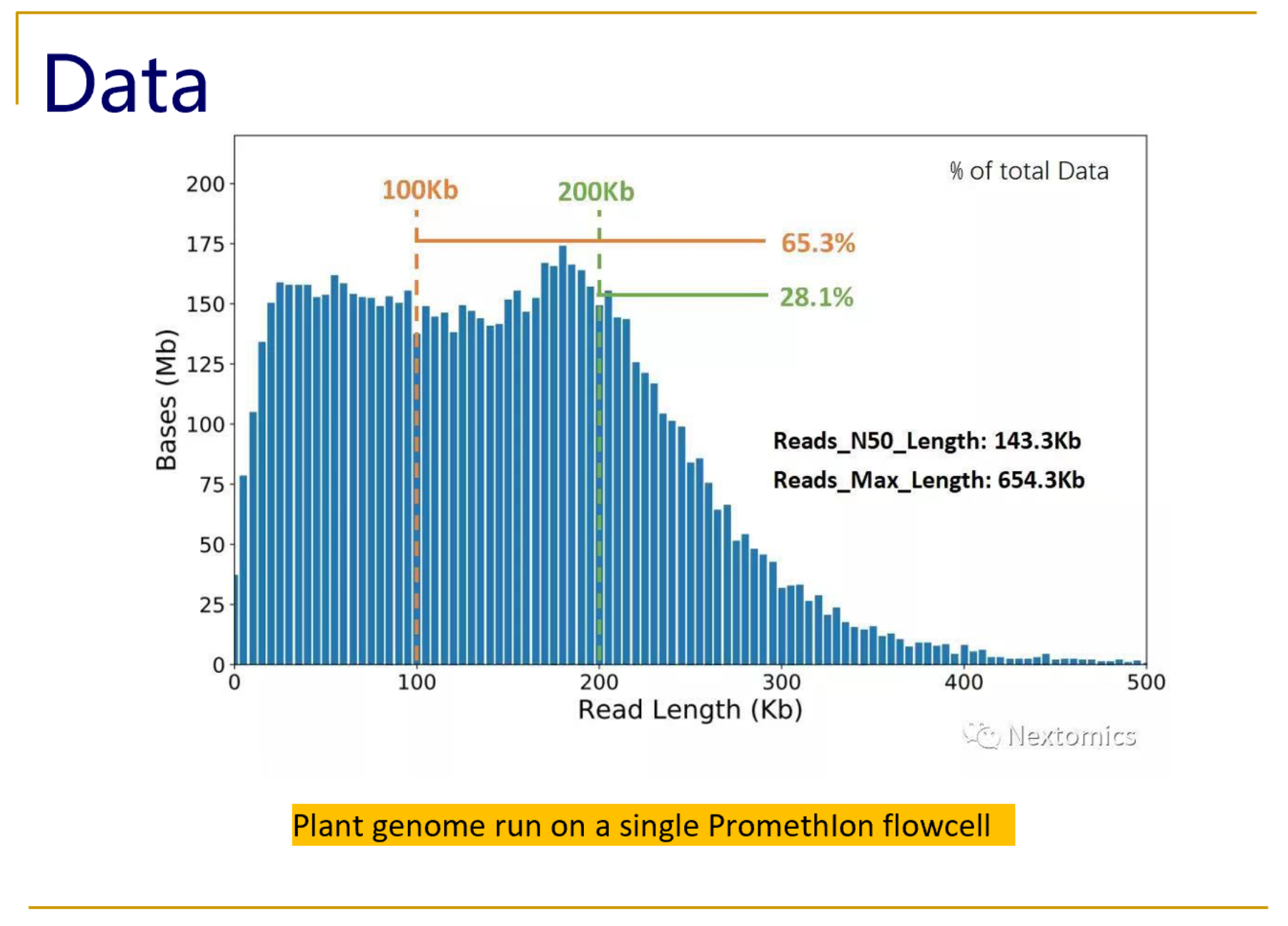
From kilobases to "whales": a short history of ultra-long reads and high-throughput genome sequencing