Boxplot of sequencing depth data and amplicons size (bp). The range of... | Download Scientific Diagram
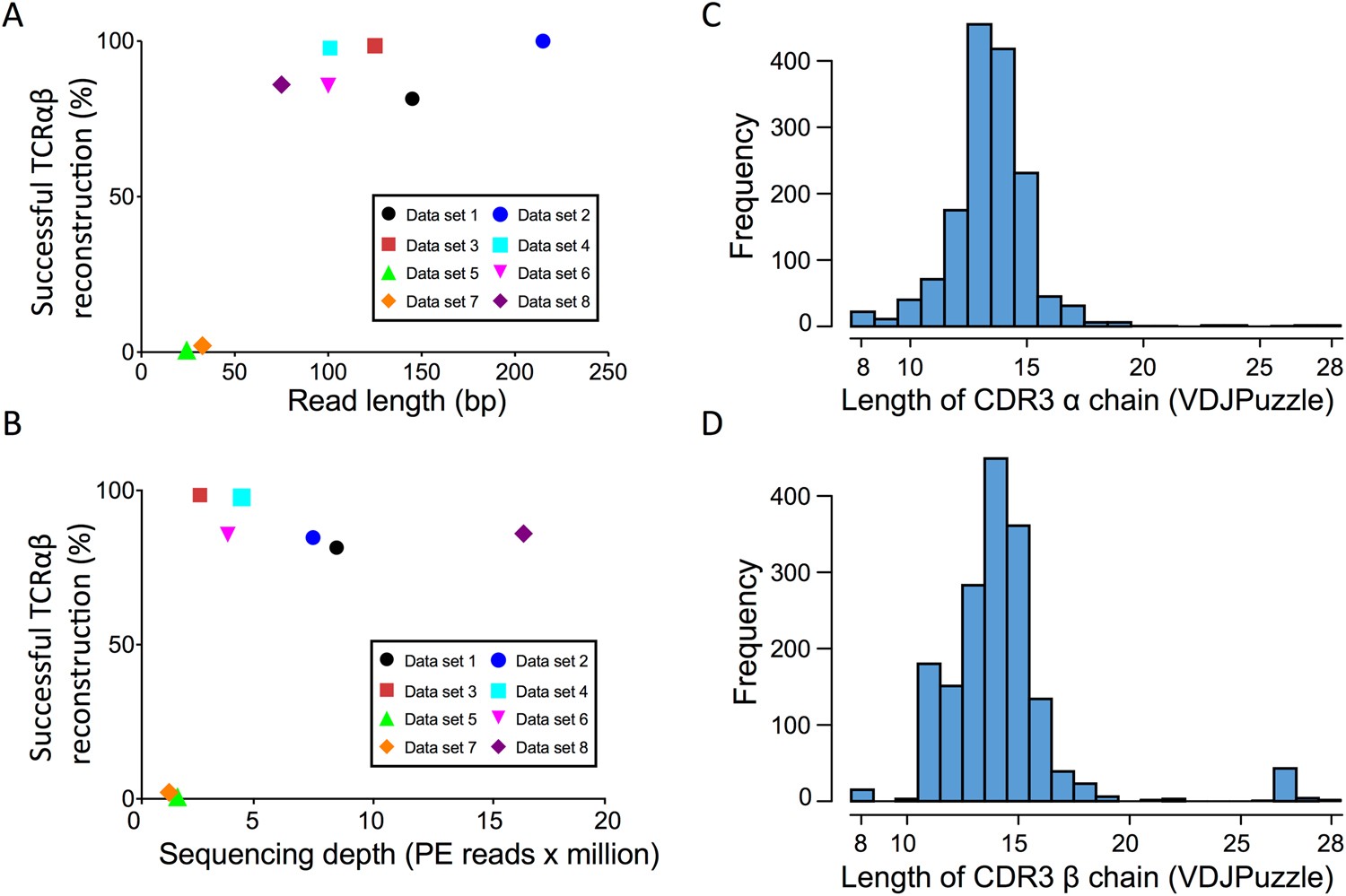
Impact of sequencing depth and read length on single cell RNA sequencing data of T cells | Scientific Reports

Recommendations for accurate genotyping of SARS-CoV-2 using amplicon-based sequencing of clinical samples - ScienceDirect

RNA Sequencing can be a bit intimidating if you're new to the platform.🤔 This is a short 📽️clip from our free "Beginner's Guide to RNA-Seq" webinar going over some decisions you need

The trade-off between increased multiplexing and decreasing sequencing depth in smallRNA-Seq | RNA-Seq Blog