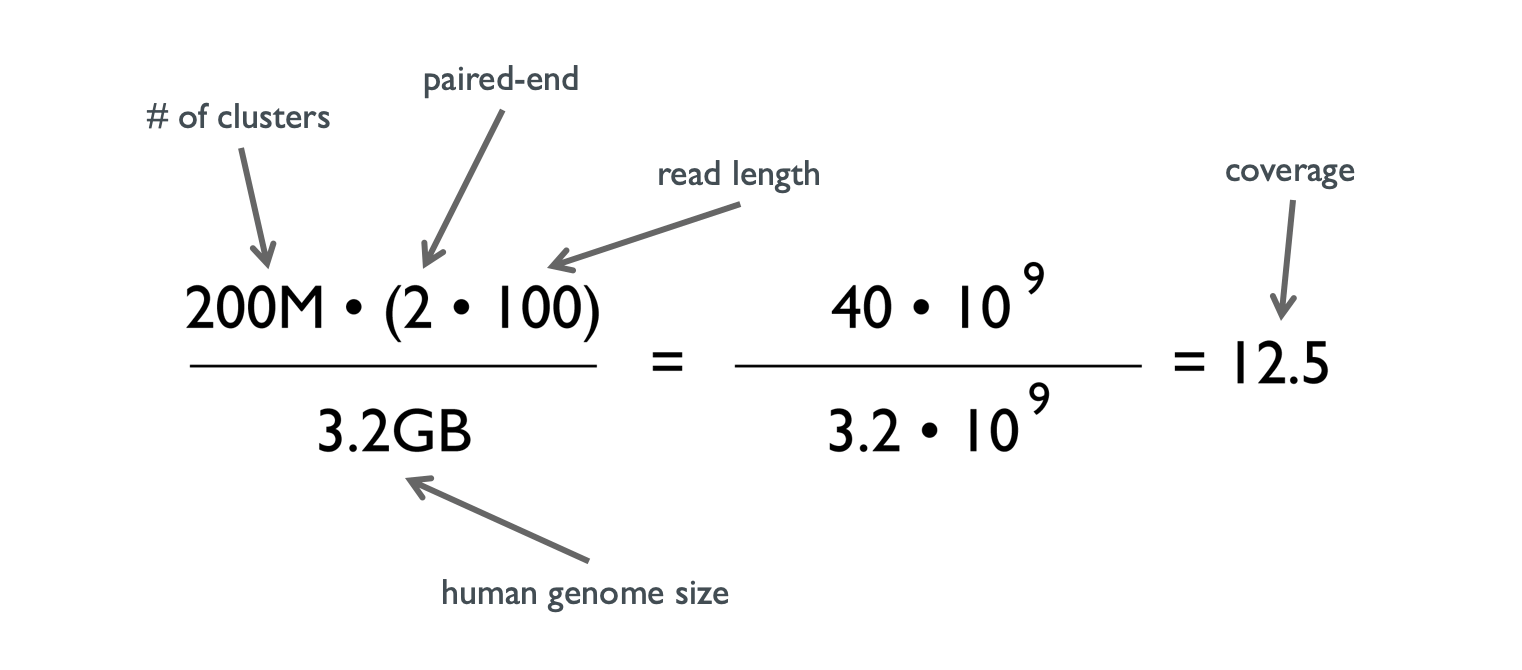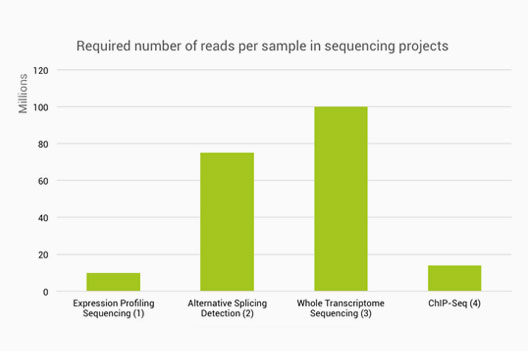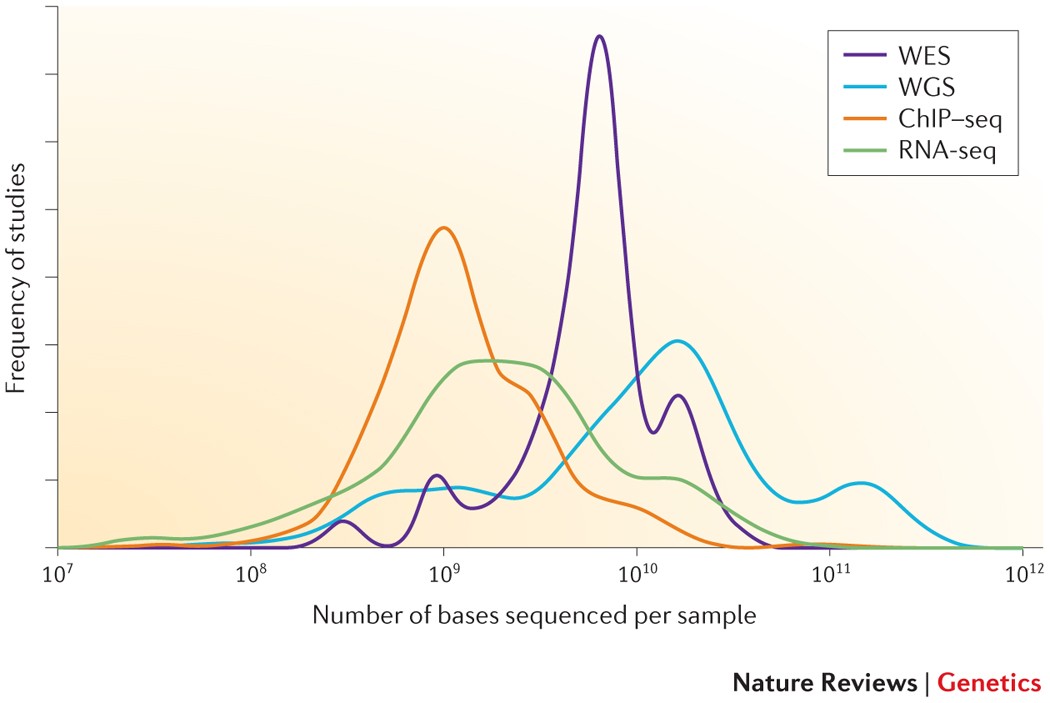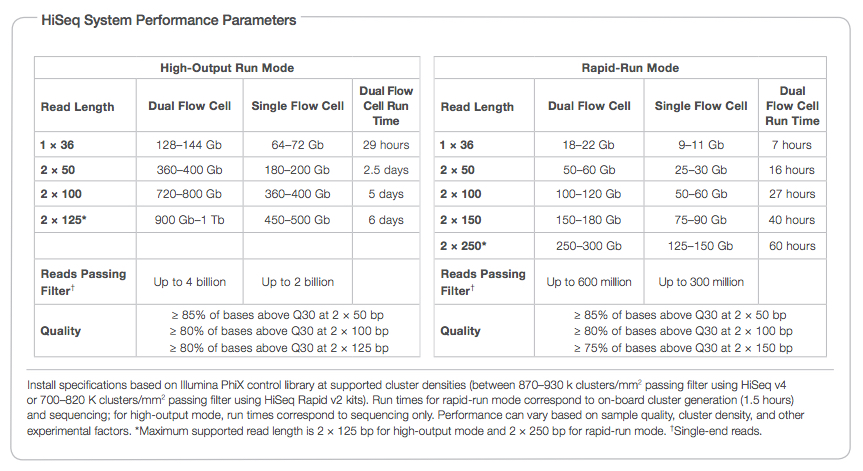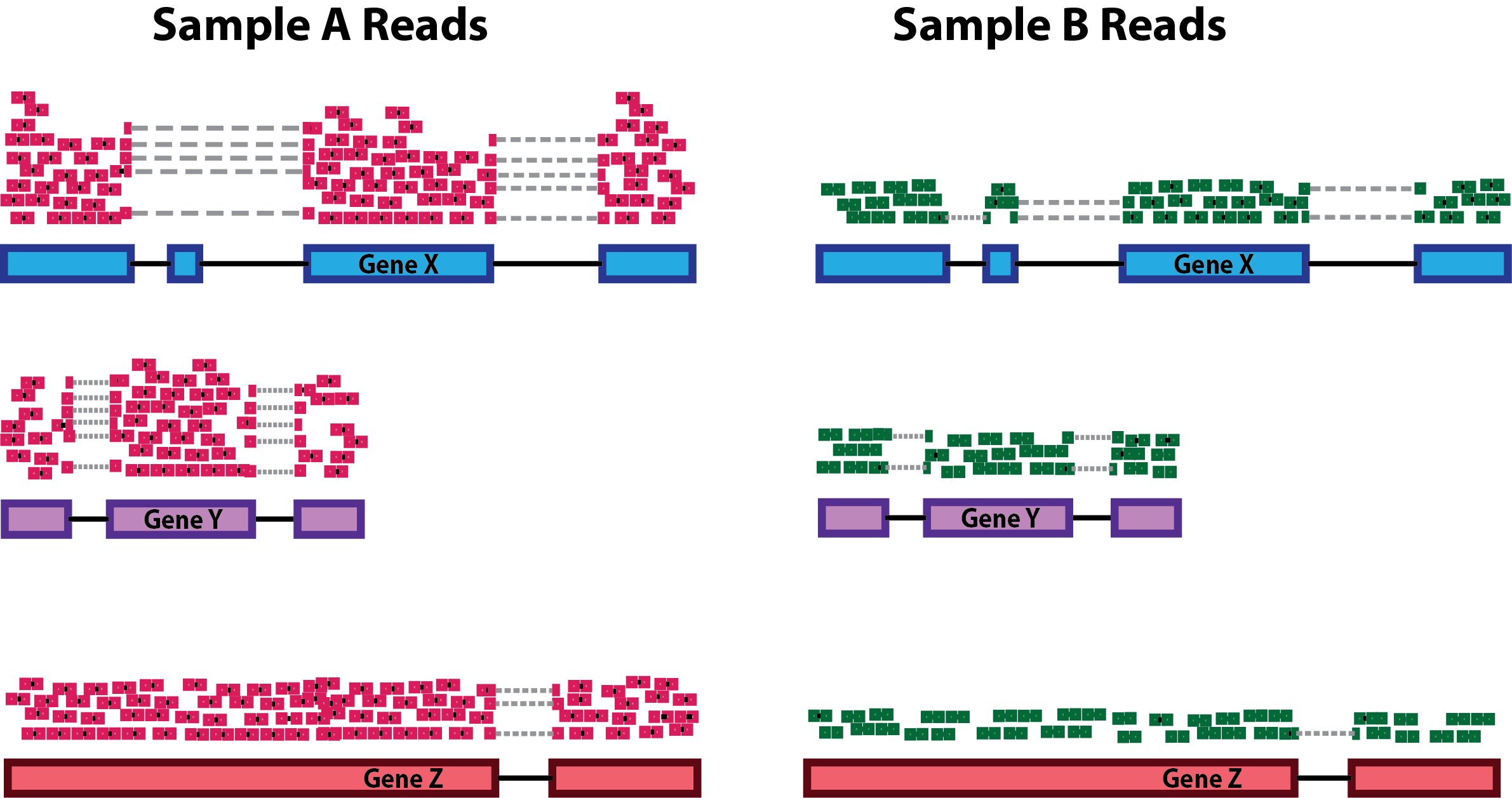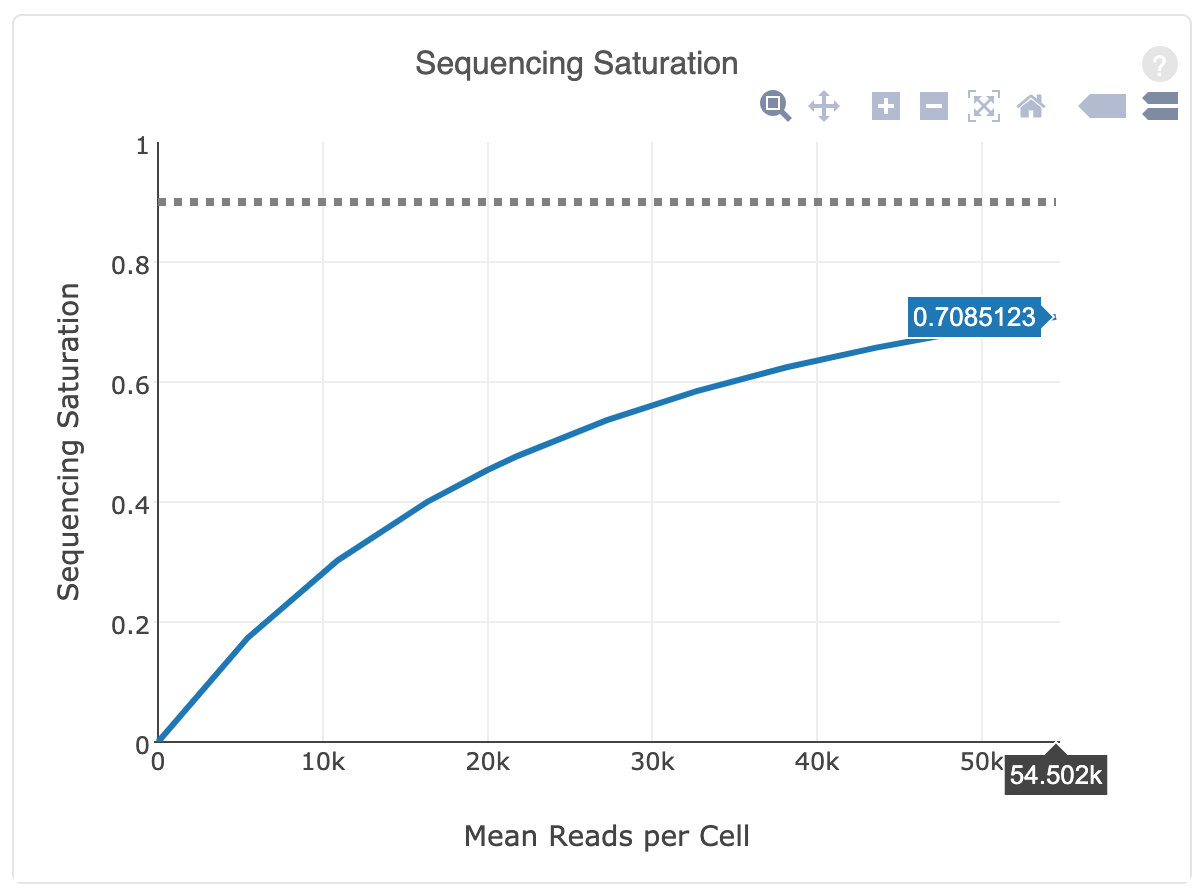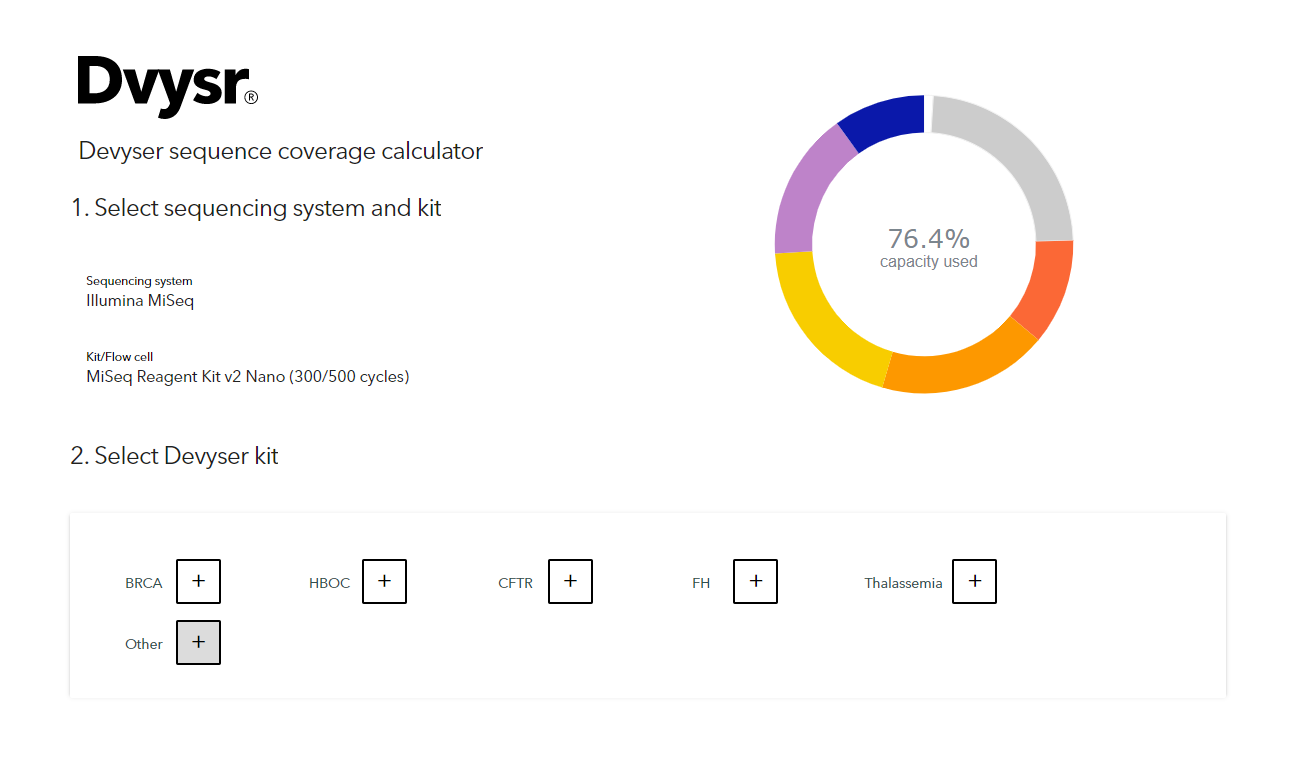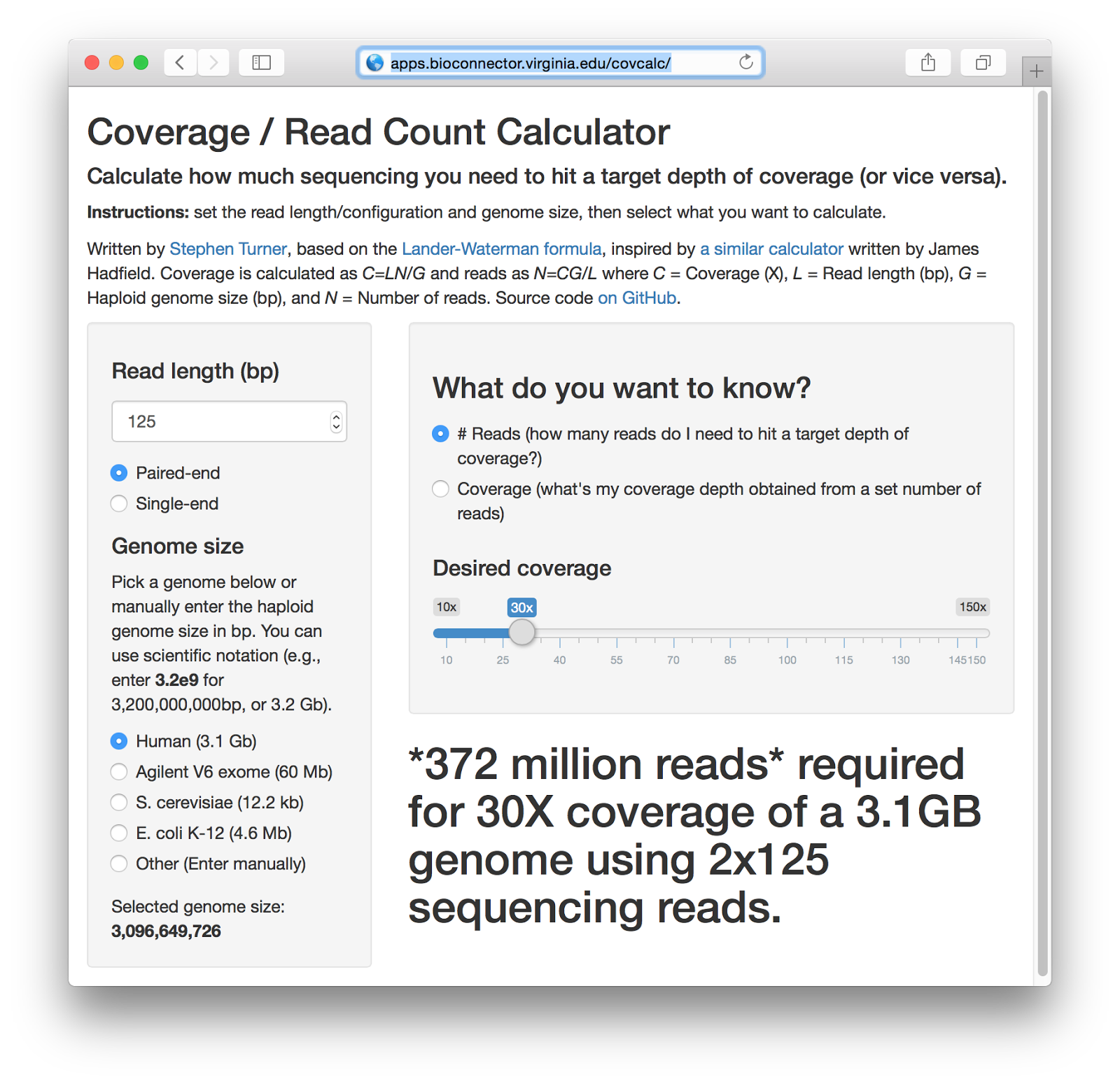
Getting Genetics Done: Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments
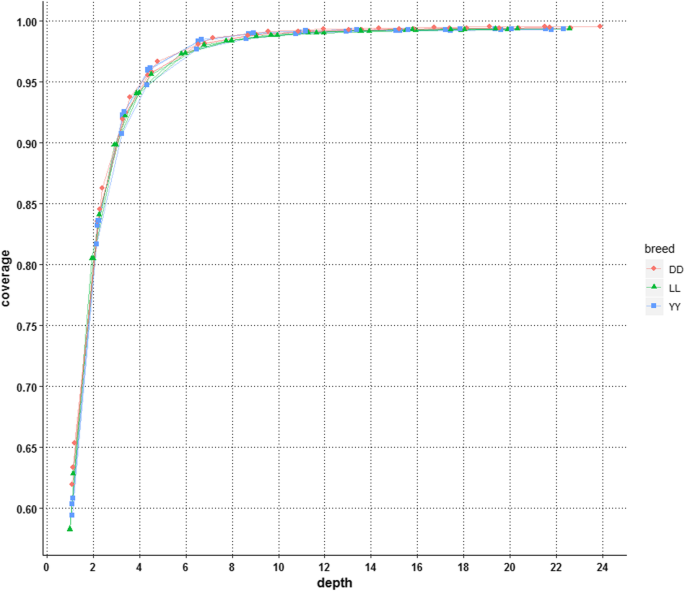
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text

Calculation of contig number for various combination of c, L and T, by... | Download Scientific Diagram

Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments | R-bloggers

Mathematical Framework Provides a Read Depth Calculator and Guidelines... | Download Scientific Diagram
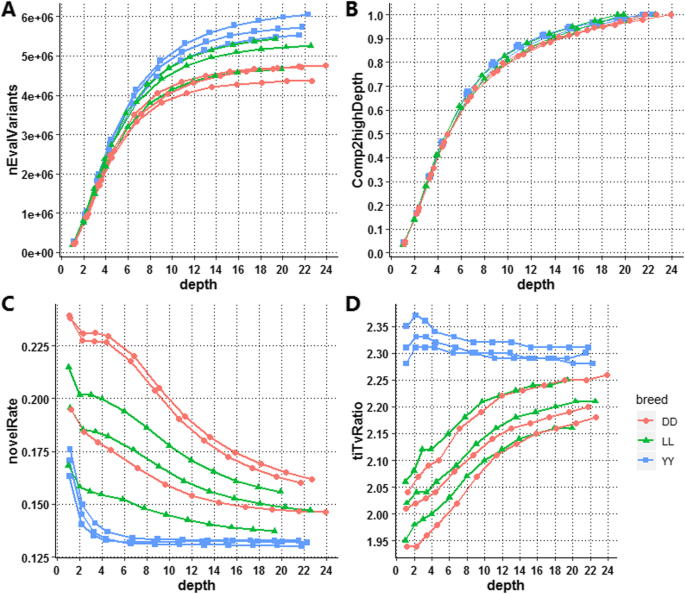
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text