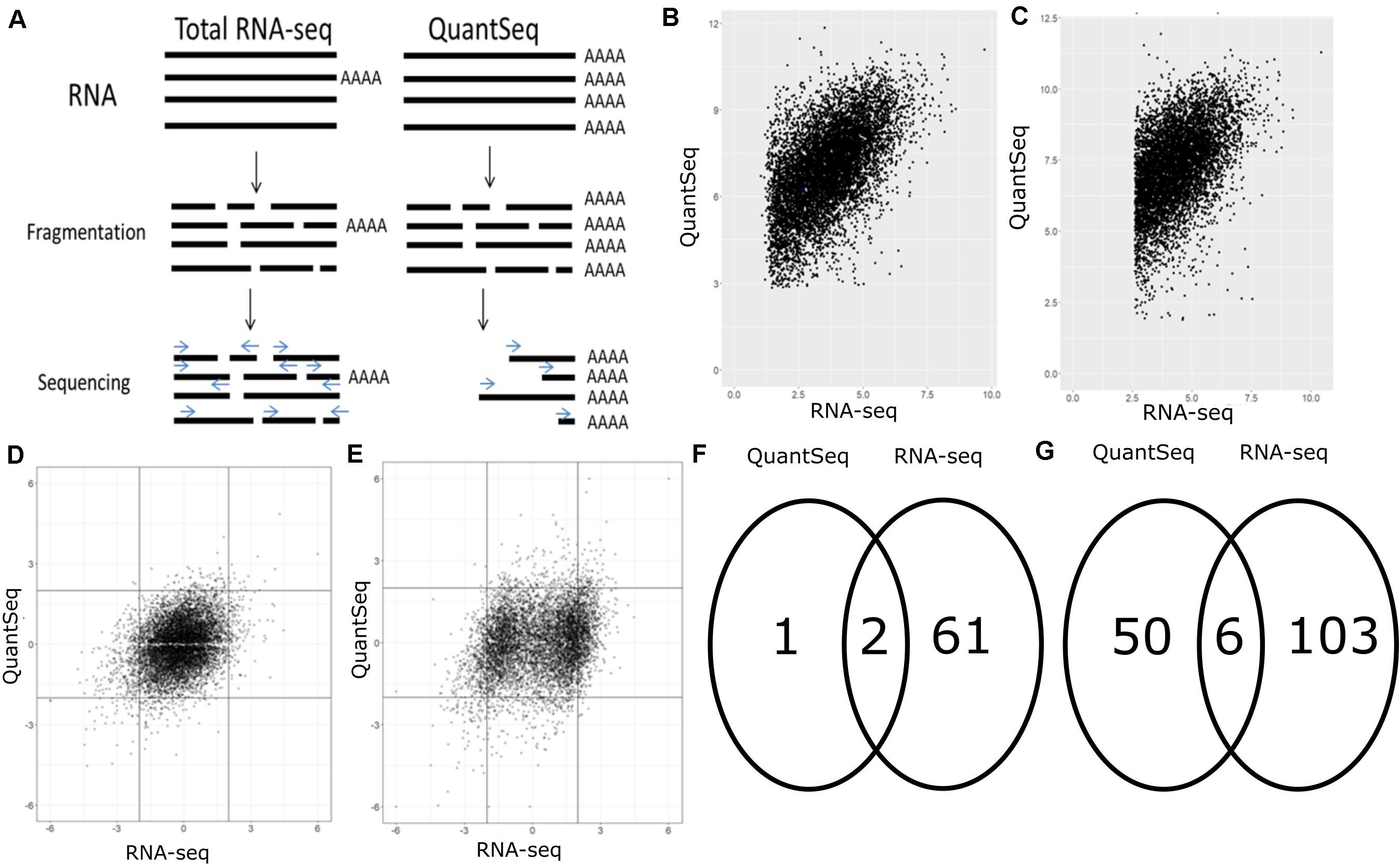
Frontiers | A Comparison of Low Read Depth QuantSeq 3′ Sequencing to Total RNA-Seq in FUS Mutant Mice
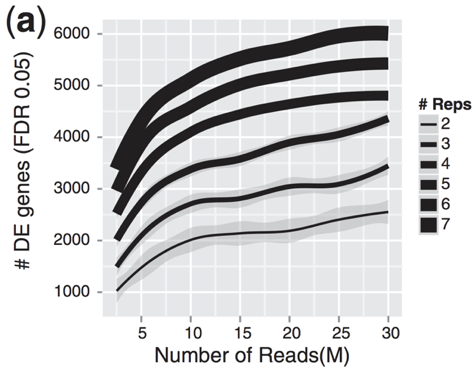
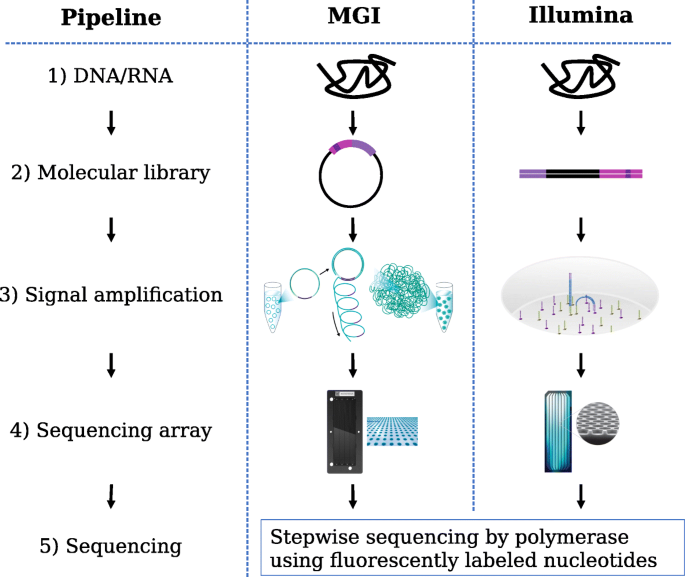
Experimental design considerations | Introduction to RNA-Seq using high-performance computing - ARCHIVED

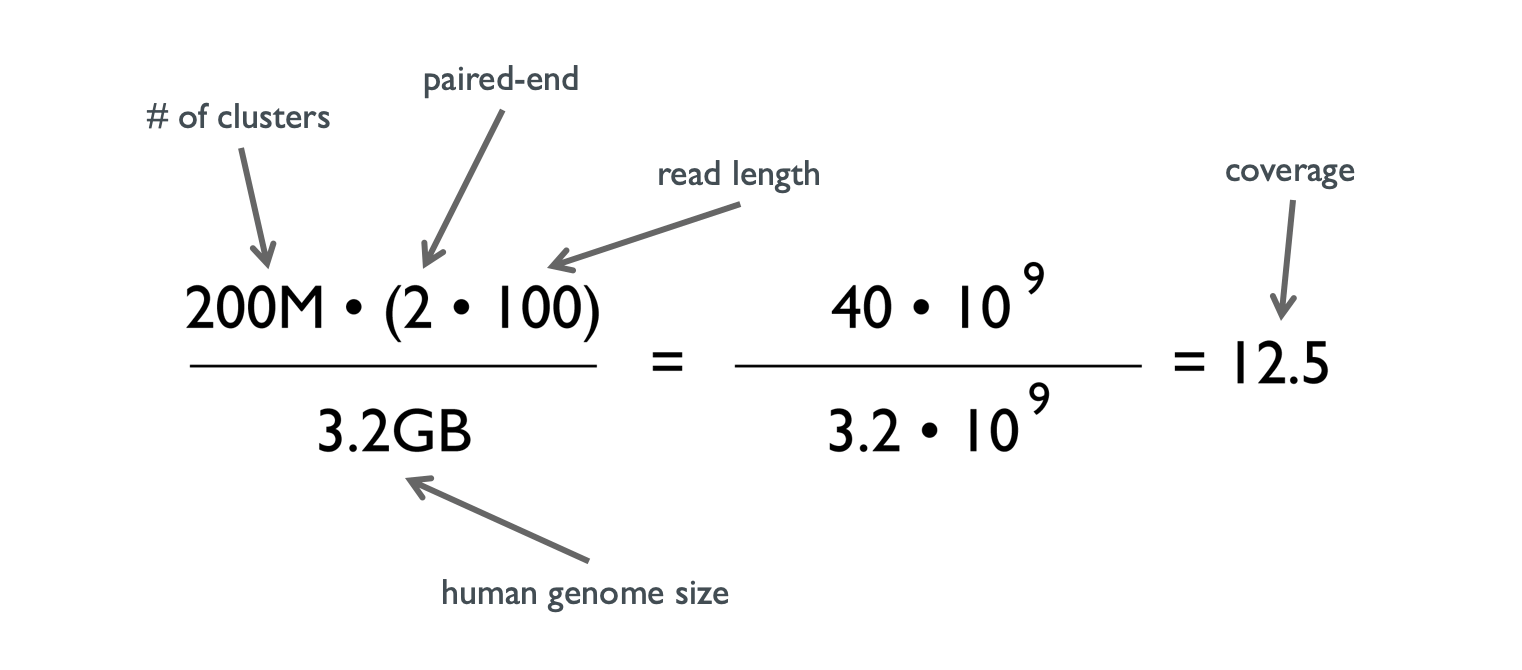
Read coverage of genome sequencing and RNA-seq data. a Read coverage of... | Download Scientific Diagram

Difference between sequencing Coverage and depth. Depth vs Coverage. Why they are important? - YouTube
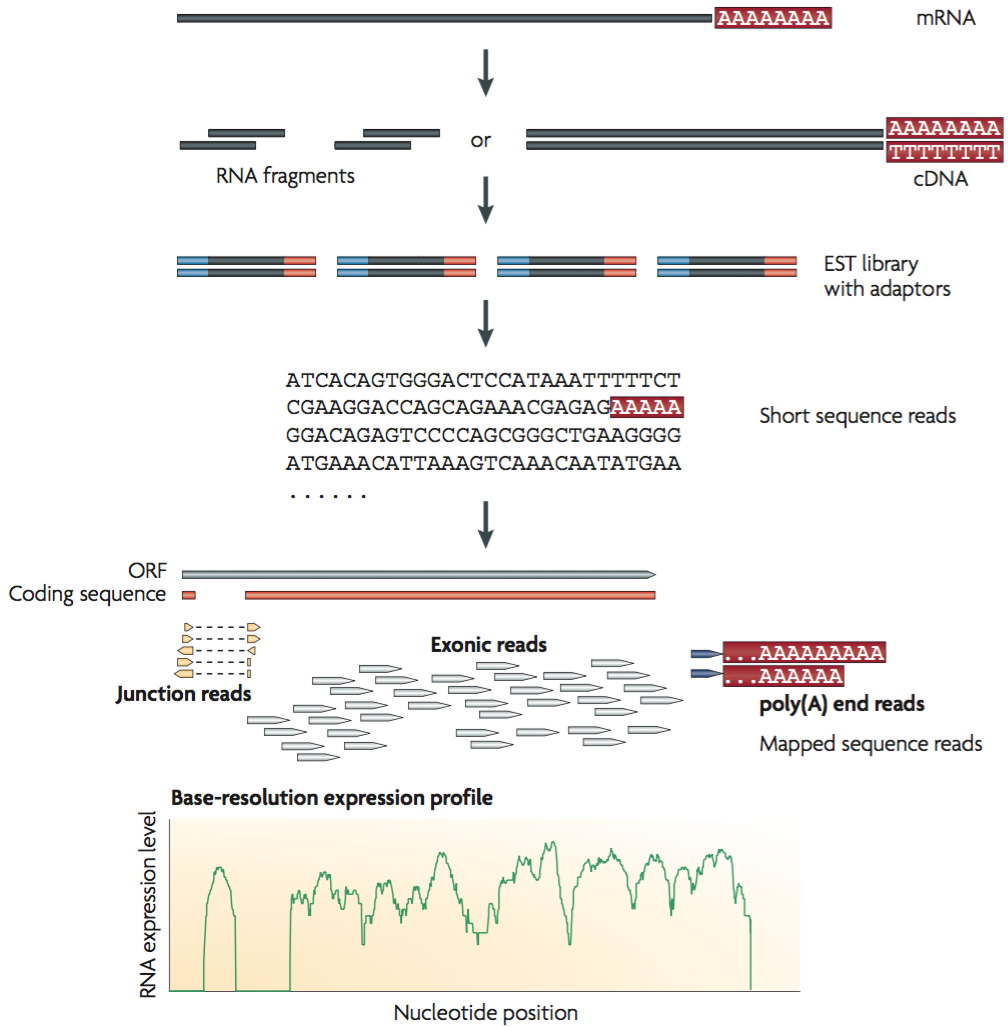
Improved Annotation of 3′ Untranslated Regions and Complex Loci by Combination of Strand-Specific Direct RNA Sequencing, RNA-Seq and ESTs | PLOS ONE

Detecting, Categorizing, and Correcting Coverage Anomalies of RNA-Seq Quantification - ScienceDirect
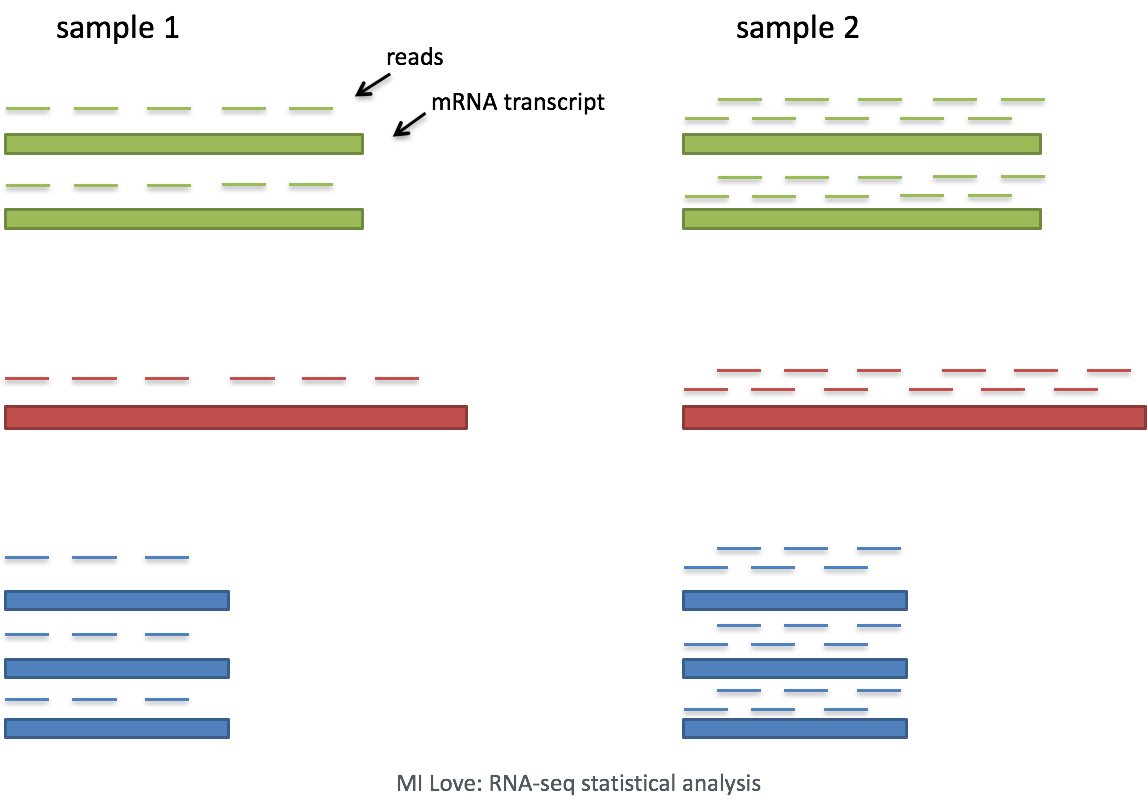
Single-cell RNA-seq: Normalization, identification of most variable genes | Introduction to single-cell RNA-seq