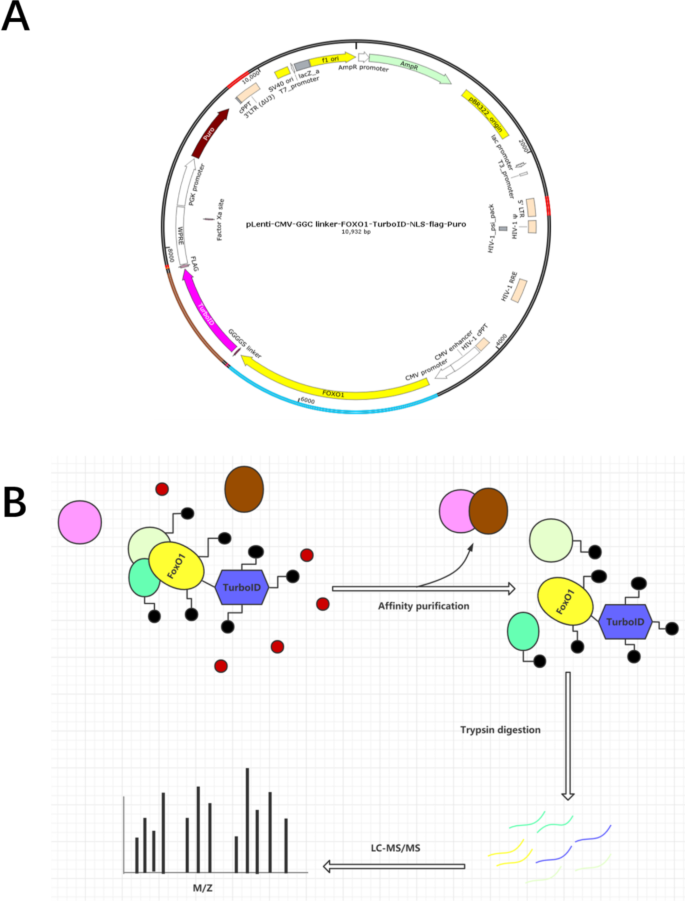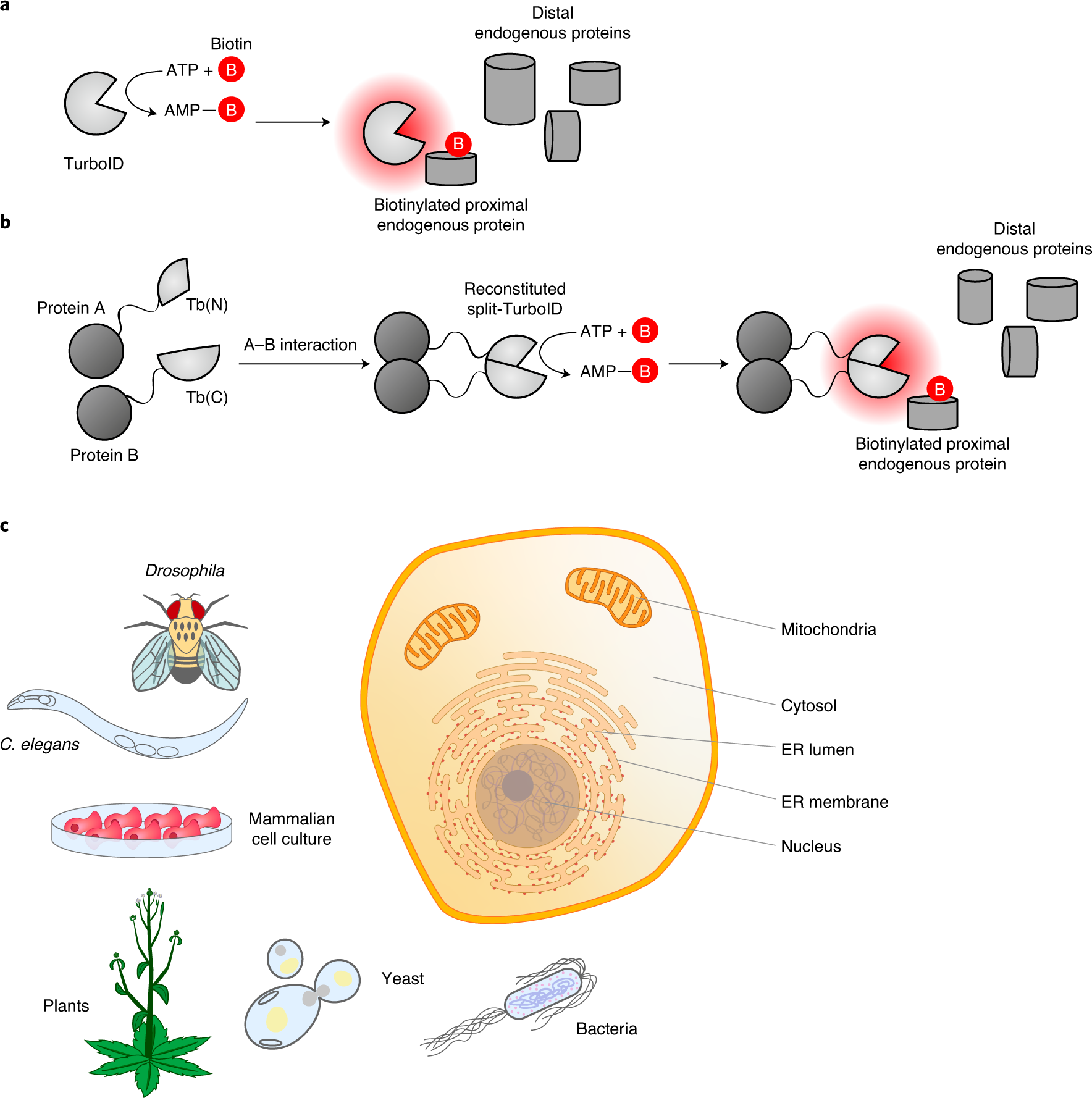
TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity | Nature Communications
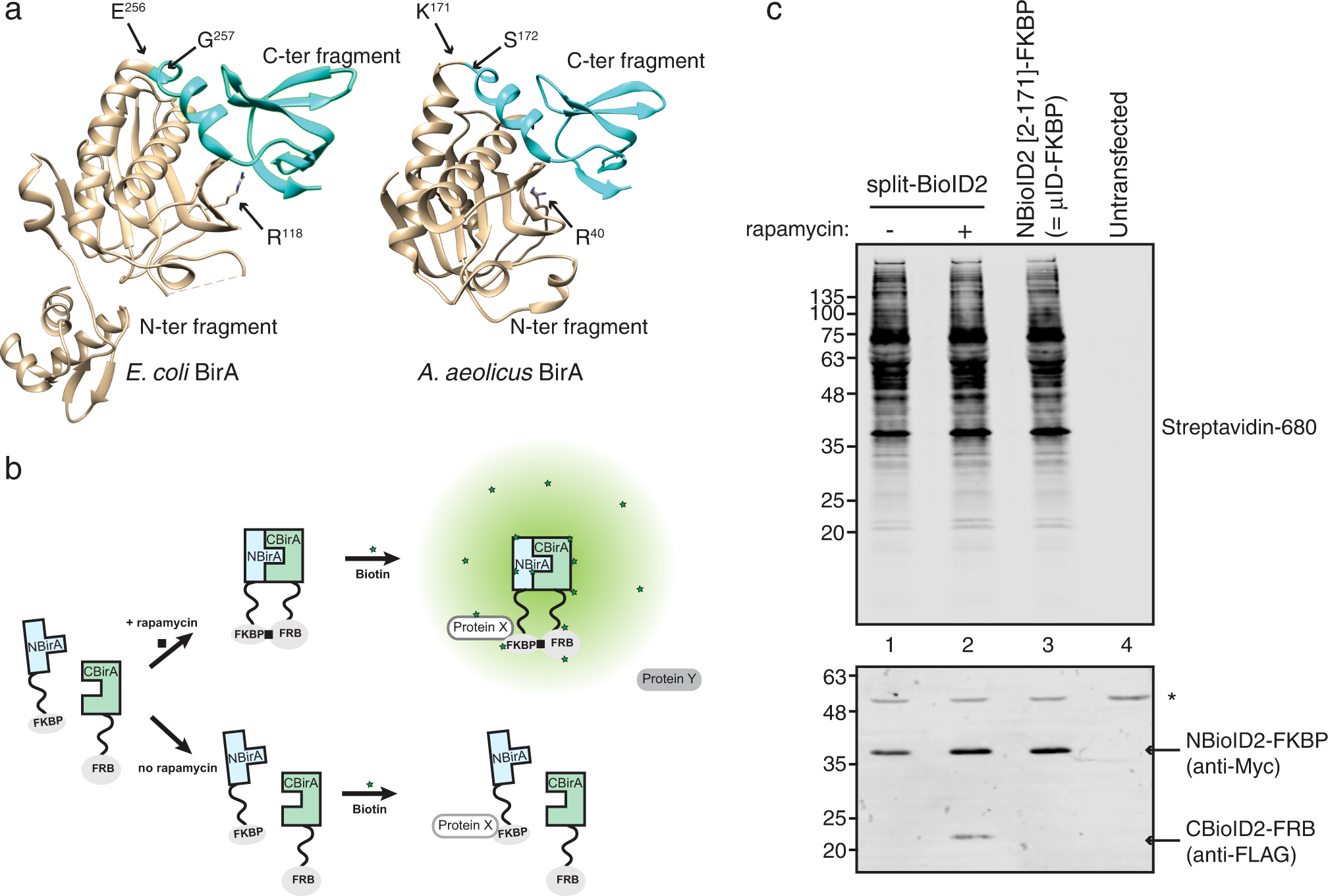
Engineering of ultraID, a compact and hyperactive enzyme for proximity-dependent biotinylation in living cells | Communications Biology

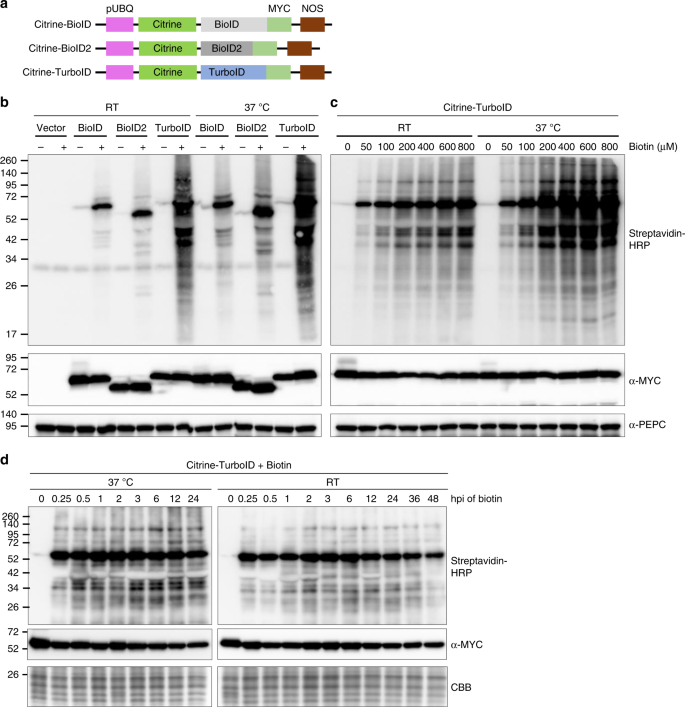
Application of TurboID-mediated proximity labeling for mapping a GSK3 kinase signaling network in Arabidopsis | bioRxiv

IJMS | Free Full-Text | Differential CFTR-Interactome Proximity Labeling Procedures Identify Enrichment in Multiple SLC Transporters

Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID | eLife

Schematic illustration of constructs used in BioID experiments. For... | Download Scientific Diagram
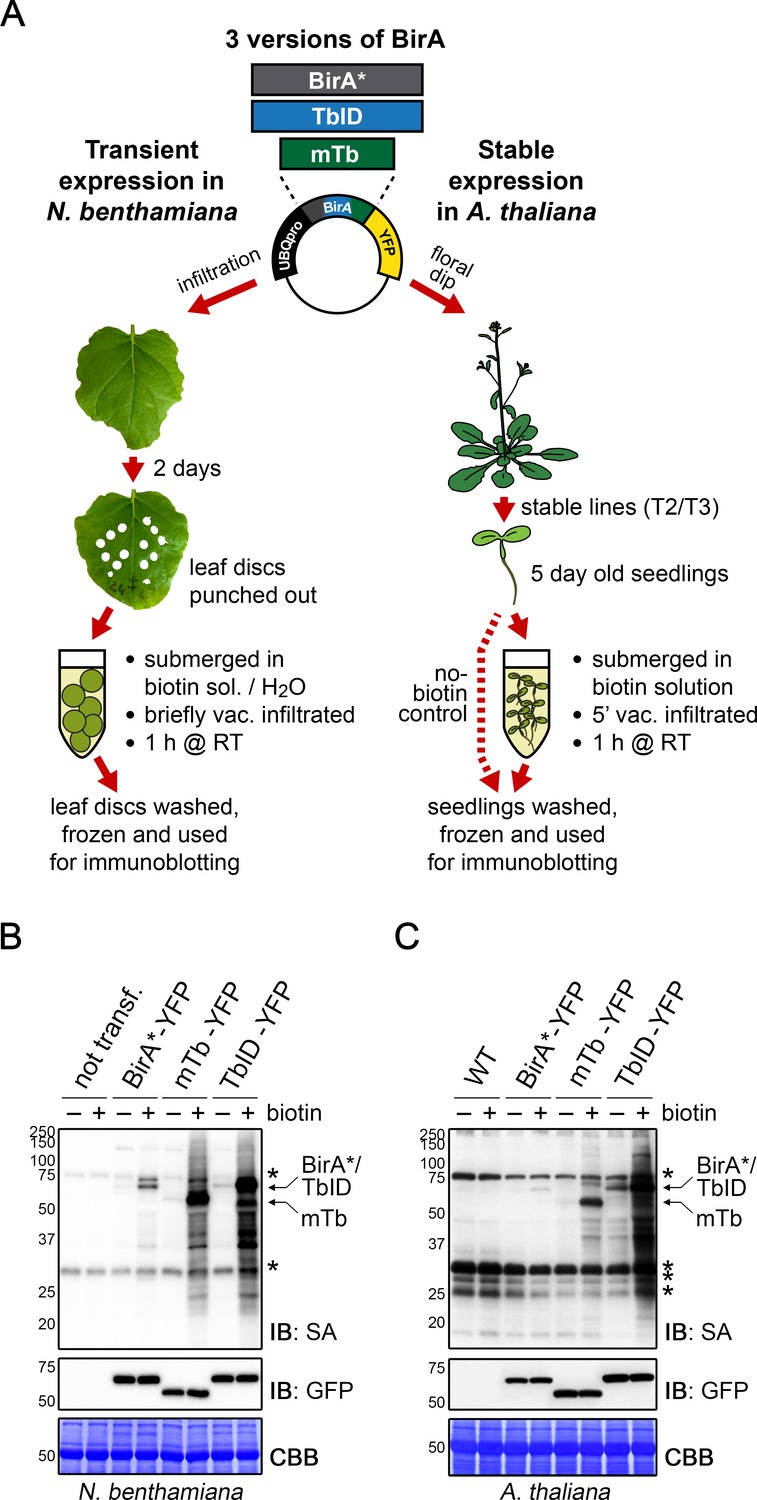
Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID | eLife

TurboID Screening of the OmpP2 Protein Reveals Host Proteins Involved in Recognition and Phagocytosis of Glaesserella parasuis by iPAM Cells | Microbiology Spectrum